| "Database" | "pubmed" | target database from which to retrieve data; options for "ESearch", "ESummary", "EFetch", "ELink", "EPost", "ESpell";
allowed values include:
"pubmed","protein","nuccore","ipg","nucleotide","structure","genome","annotinfo","assembly","bioproject",
"biosample","blastdbinfo","books","cdd","clinvar","gap","gapplus","grasp","dbvar","gene","gds","geoprofiles",
"medgen","mesh","nlmcatalog","omim","orgtrack","pmc","proteinclusters","pcassay","protfam","pccompound",
"pcsubstance","seqannot","snp","sra","taxonomy","biocollections","gtr" |
| "RetMode" | "Dataset" or
"Text" | format of the returned output; options for "ESummary", "EFetch", "ELink"; allowed values include:
"Dataset", "Text", "XML"; For "EFetch", some databases, including "bioproject", "clinvar", "taxonomy", "snp", "sra",
and "gtr", return "Dataset" instead of "Text" by default;
For "EFetch", some databases, including "nuccore" and "nucleotide", "BioSequence" output is also supported |
| "RetType" | None | type of retrived data; options for "EFetch"; allowed values vary by database:
all databases: "docsum", "uilist"
gene: "gene_table"
protein: "acc", "fasta", "seqid"
nuccore, nucleotide: "acc", "seqid", "fasta", "fasta_cds_na", "fasta_cds_aa"
pmc: "medline"
pubmed: "medline", "abstract" |
| "UseHistory" | False | posting the UIDs resulting from the search operation onto the NCBI History server; options for "ESearch";
"UseHistory" must be set to True for "ESearch" to accept a WebEnv as input |
| "WebEnv" | None | web environment string returned from a previous "ESearch", "EPost" or "ELink" call; when provided, NCBIEntrezData will append the results to the existing environment; "UseHistory" must be set to True for "ESearch" to accept a WebEnv as input;
options for "ESearch", "ESummary", "EFetch" |
| "QueryKey" | None | query key returned by a previous ESearch, EPost or ELink call; when provided, NCBIEntrezData will find the intersection of the set specified by QueryKey and the set retrieved by the query; options for "ESearch", "ESummary", "EFetch" |
| "RetStart" | 0 | sequential index of the first UID in the retrieved set to be shown in the output (default=0, corresponding to the first record of the entire set);
options for "ESearch" |
| "RetMax" | 20 | total number of UIDs from the retrieved set to be shown in the output; options for "ESearch" |
| "Sort" | None | method used to sort UIDs in the output; options for "ESearch";
allowed values vary by database and include:
gene: "relevance", "name"
pubmed: "pub_date", "Author", "JournalName", "relevance" |
| "Field" | None | search field to limit a result of "ESearch"; allowed values vary by database and are found in the result of NCBIEntrezData[database, "EInfo"] |
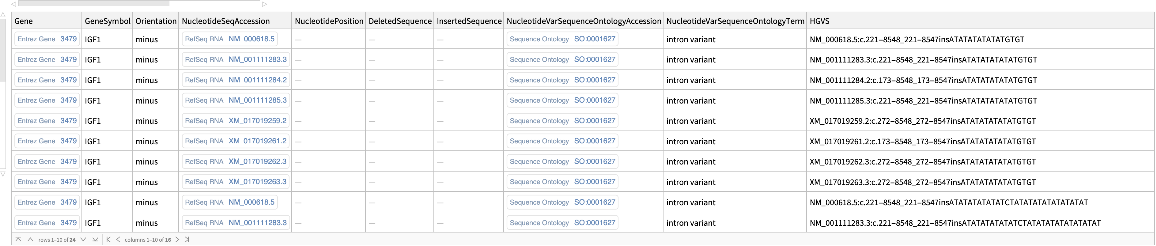
| "IDType" | None | type of identifier to return for sequence databases (nuccore, protein); if not specified, GenInfo Identifier (GI) numbers are returned; other value includes "acc", returning sequence accession numbers instead;
options for "ESearch", "ELink" |
| "DateType" | None | type of date used to limit a search; options for "ESearch", "ELink";
allowed values include:
"mdat" (modification date), "pdat" (publication date), "edat" (Entrez date) |
| "RelDate" | None | integer n limiting the result that has a date specified by "DateType" within the last n days; options for "ESearch", "ELink" |
| "MinDate" | None | date range used to limit a search result of "ESearch" or "ELink" by the date specified by "DateType"; two parameters, "MinDate" and "MaxDate", must be used together to specify an arbitrary date range; the general date format is YYYY/MM/DD, and these variants are also allowed: YYYY, YYYY/MM |
| "MaxDate" | None | date range used to limit a search result of "ESearch" or "ELink" by the date specified by "DateType"; two parameters, "MinDate" and "MaxDate", must be used together to specify an arbitrary date range; the general date format is YYYY/MM/DD, and these variants are also allowed: YYYY, YYYY/MM |
| "Strand" | "plus" | strand of DNA to retrieve for "EFetch"; allowed values are "plus" or "minus" |
| "SeqStart" | None | integer coordinate of the first sequence base to retrieve; options for "EFetch" |
| "SeqStop" | None | integer coordinate of the last sequence base to retrieve; options for "EFetch" |
| "Complexity" | None | data content to return, specified by integer; many sequence records are part of a larger data structure or "blob", and the complexity parameter determines how much of that blob to return; options for "EFetch"; allowed values include:
0 (entire blob), 1(bioseq), 2 (minimal bioseq-set), 3 (minimal nuc-prot), 4 (minimal pub-set) |
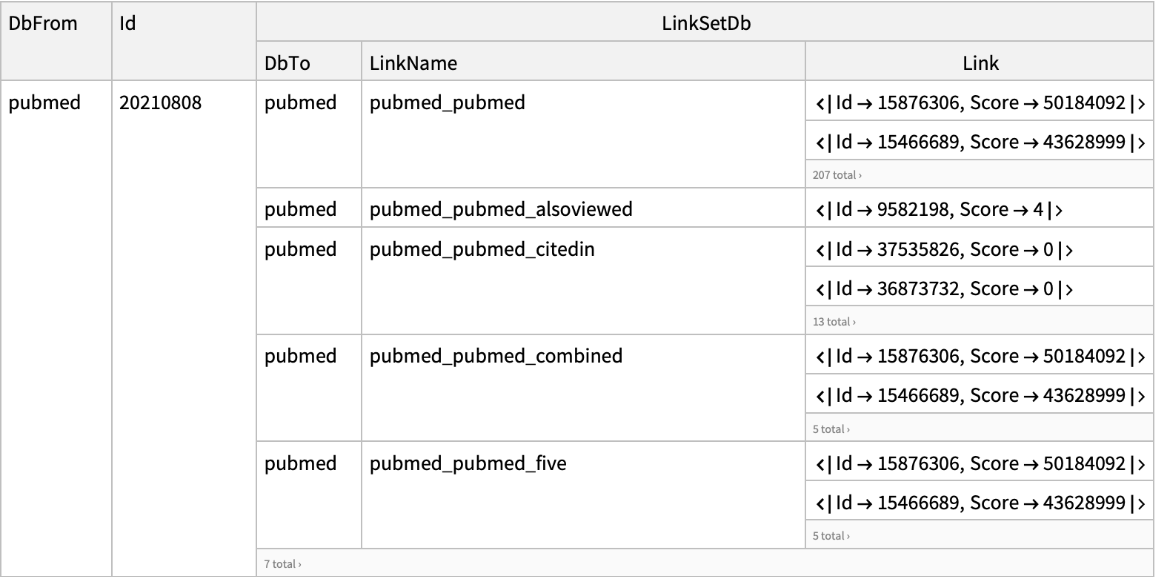
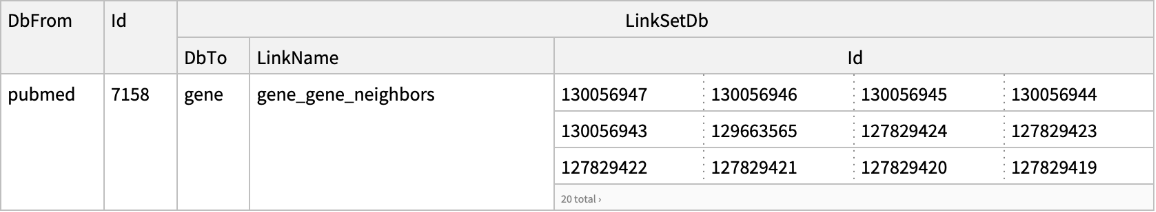
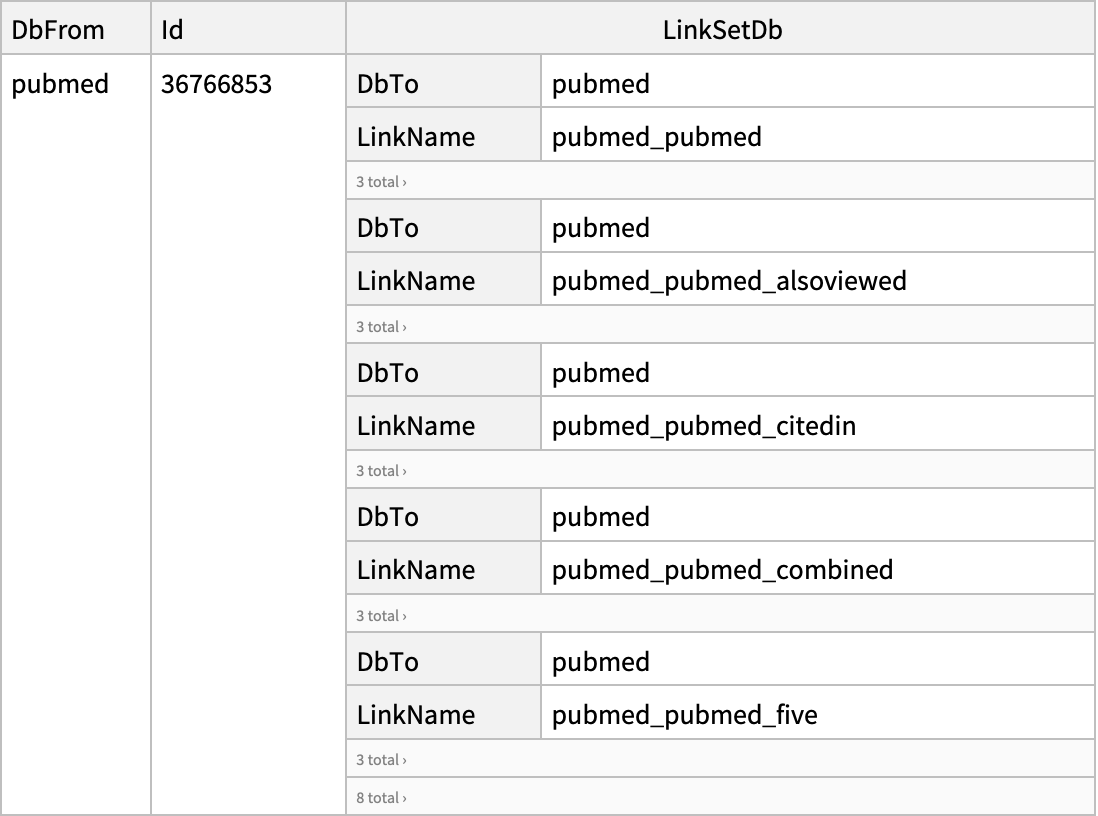
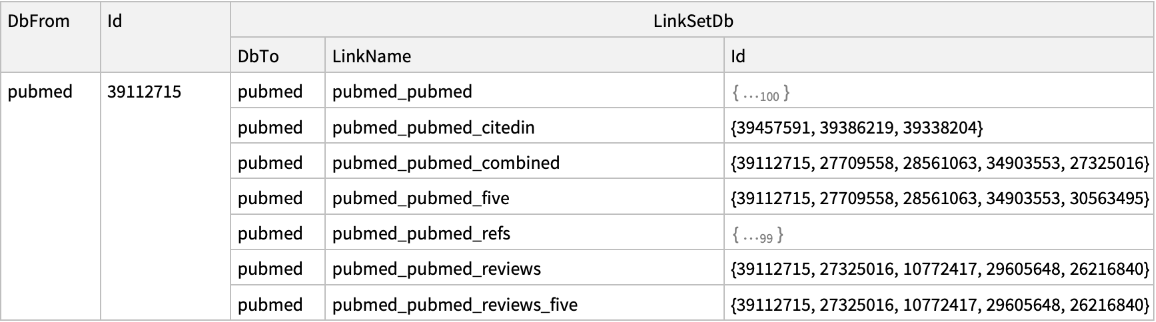
| "DatabaseFrom" | "pubmed" | name of the database containing the input UIDs; options for "ELink"; if "Database" and "DatabaseFrom" are set to the same database value,
then ELink will return computational neighbors within that database; for available computational neighbors see the full list of Entrez links |
| "CommandMode" | "neighbor" | command mode specified which function "ELink" will perform; allowed values include:
"neighbor": returns a set of UIDs in the "Database" linked to the input UIDs in the "DatabaseFrom"
"neighbor_score": returns a set of UIDs in a database that are similar or related to the input UIDs, along with the the computed similarity scores "neighbor_history": functions similarly to neighbor, but stores the results to the NCBI History server and returns a Web environment string and query keys for each set of resulting UIDs
"acheck": returns a list of all of the available links for the input UIDs
"ncheck": checks for the existence of links between the input UIDs and other UIDs in the same database
"lcheck": checks for the existence of external links for the set of input UIDs
"llinks": checks for the existence of external links for the set of input UIDs, and returns URLs and provider attributes for all non-library providers
"llinkslib": checks for the existence of external links for the set of input UIDs, and returns URLs and provider attributes for all providers
"prlinks": returns the primary external link provider for each input UID |
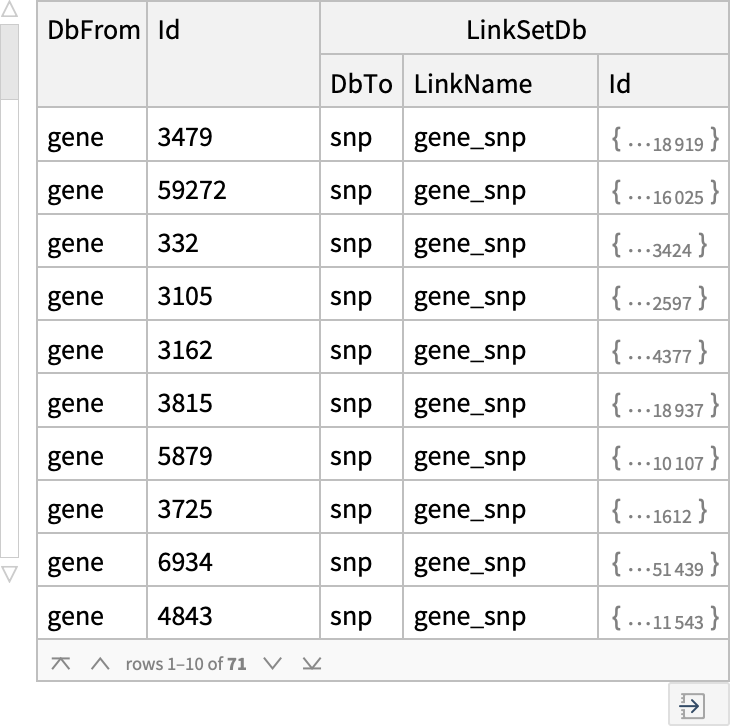
| "LinkName" | None | name of the Entrez link used for "ELink" to retrieve; see the full list of Entrez links |
| "Term" | None | string term used to limit the output set of "ELink" UIDs; the "Term" parameter only functions when "Database" and "DatabaseFrom" are set to the same database value |
| "Holding" | None | string name of the external link provide for "ELink"; the "Holding" parameter only functions when "CommandMode" is set to "llinks" or "llinkslib" |
| APIKey | None | an API key provided by NCBI; by including a key, up to 10 requests per second are permitted by default |


![ResourceFunction["NCBIEntrezData"][
ExternalIdentifier["RefSeqGenomeAccessionNumber", "5"], "EFetch", {"Database" -> "nucleotide", "RetMode" -> "BioSequence"}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/68f93290414f6230.png)
![ResourceFunction["NCBIEntrezData"][
ExternalIdentifier["RefSeqGenomeAccessionNumber", "5"], "EFetch", {"Database" -> "nucleotide", "RetType" -> "fasta_cds_na", "RetMode" -> "BioSequence"}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/237ddf6295a1e895.png)
![ResourceFunction["NCBIEntrezData"][
ExternalIdentifier["RefSeqGenomeAccessionNumber", "5"], "EFetch", {"Database" -> "nucleotide", "RetType" -> "fasta_cds_aa", "RetMode" -> "BioSequence"}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/36f8482c9e3b4e43.png)

![cancerpost = ResourceFunction[
"NCBIEntrezData"][{"40048260", "40048237", "40048221", "40048212", "40048195", "40048190", "40048187", "40048163", "40048159", "40048143", "40048083", "40048052", "40048045", "40048042", "40048041", "40048036", "40048034", "40048031", "40048030", "40048028"}, "EPost"]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/15bae896c797f398.png)

![ResourceFunction["NCBIEntrezData"]["lung cancer", "ESearch", "UseHistory" -> True, "QueryKey" -> cancerpost["QueryKey"], "WebEnv" -> cancerpost["WebEnv"]]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/2993e79163eec5b7.png)




![ResourceFunction[
"NCBIEntrezData"]["Alzheimer's disease treatment", "ESearch", {"UseHistory" -> True, "WebEnv" -> "MCID_67ca330f0575e56b50054e34"}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/38bbd267c3119172.png)

![ResourceFunction[
"NCBIEntrezData"]["genetic variation", "ESearch", {"UseHistory" -> True, "WebEnv" -> "MCID_67ca330f0575e56b50054e34", "QueryKey" -> 2}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/334ff1ea5aaf4768.png)




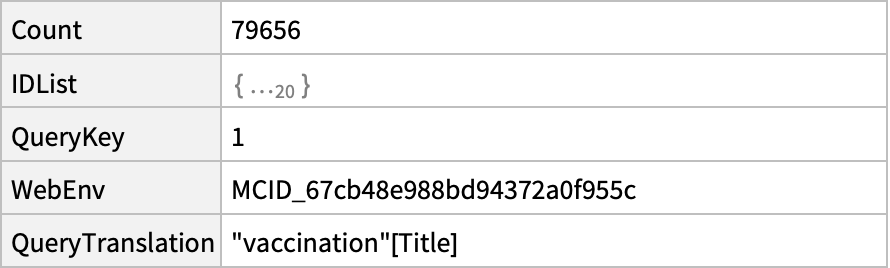
![ResourceFunction[
"NCBIEntrezData"]["mitogen activated protein kinase", "ESearch", {"Database" -> "protein", "IDType" -> "acc"}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/6c916a1017d34844.png)
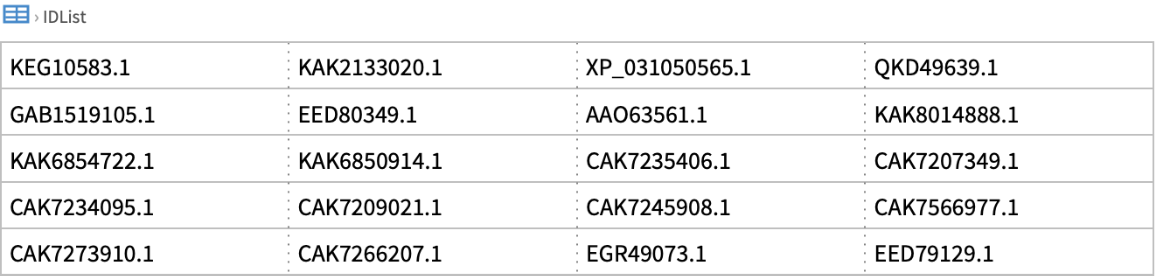
![ResourceFunction[
"NCBIEntrezData"]["signal transduction", "ESearch", {"MinDate" -> DateObject[{2025, 1, 1}, "Day"], "MaxDate" -> DateObject[{2025, 1, 31}, "Day"], "DateType" -> "pdat"}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/5d3938cd8748da3a.png)


![ResourceFunction[
"NCBIEntrezData"]["signal transduction", "ESearch", {"DateType" -> "pdat", "MinDate" -> DateObject[{2024, 1}, "Month"], "MaxDate" -> DateObject[{2024, 12}, "Month"]}]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/18bd4b129ca034bc.png)




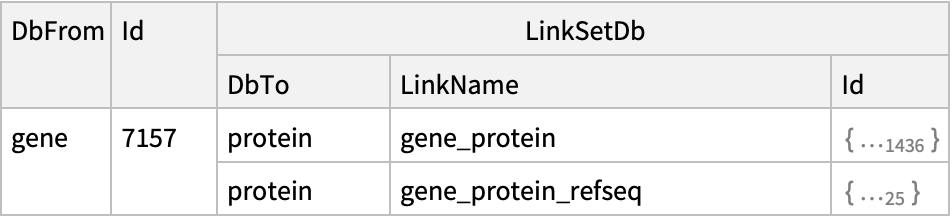
![ResourceFunction[
"NCBIEntrezData"]["Severe acute respiratory syndrome coronavirus 2[Organism]", "ESearch", "Database" -> "nucleotide", "IDType" -> "acc"]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/472a277df696e495.png)

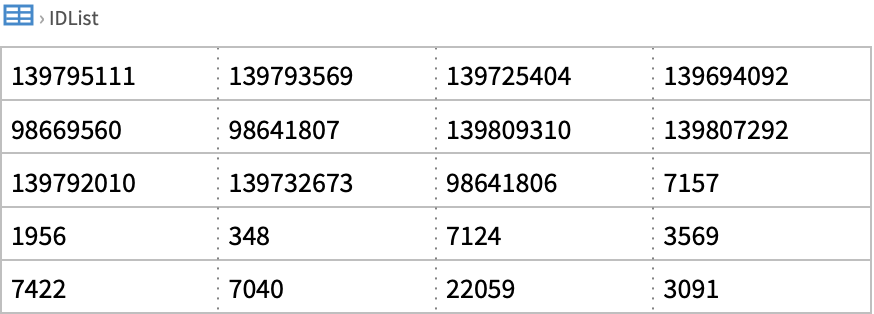
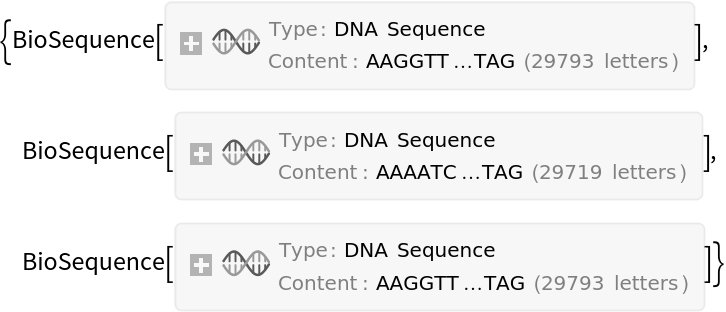
![ids = ResourceFunction["NCBIEntrezData"][
"Severe acute respiratory syndrome coronavirus 2[Organism]", "ESearch", "Database" -> "nucleotide", "IDType" -> "acc", "RetStart" -> 10000, "RetMax" -> 30]["IDList"]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/42cb5f658592fb4b.png)
![pancreaticislet = ResourceFunction["NCBIEntrezData"]["Homo sapiens pancreatic islet", "ESearch", "Database" -> "gene", "RetMax" -> 100]](https://www.wolframcloud.com/obj/resourcesystem/images/d80/d80a7c19-f41d-4eed-bc48-fa2db62656e0/496f377c694b3e5a.png)