Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Retrieve features that overlap a given genomic region
ResourceFunction["EnsemblGenomeRegion"][chromosome,{start,end}] retrieve genomic features that overlap a given chromosome region specified by the start and end positions and return information in a Dataset format. | |
ResourceFunction["EnsemblGenomeRegion"][chromosome,{start,end},format] retrieve genomic sequence that overlap a given chromosome region specified by the start and end positions in a specified format. |
| "Dataset" | genomic features returned as a Dataset (default) |
| "BioSequence" | genomic sequence returned as a BioSequence object; applicable to the "sequence" Feature |
| "FASTA" | genomic sequence returned in FASTA format; applicable to the "sequence" Feature |
| "Species" | "Homo sapiens " | species for which to query; genome assemblies of following species are supported: Homo sapiens (human), Mus musculus (mouse), Danio rerio (zebrafish), Caenorhabditis elegans (nematode), Saccharomyces cerevisiae (yeast) |
| "Assembly" | None | assembly version for which to query sequence; if not specified the latest available version is used |
| "Mask" | None | option to query the sequence masked for repeat sequences; "Hard" will mask all repeats as N's and "Soft" will mask repeats as lower cased characters |
| "Strand" | 1 | strand of the nucleotide sequence to retrieve; allowed values are 1 or -1 |
| "Feature" | “sequence" | type of genomic feature to retrieve; list of multiple values are also accepted; allowed values include: "sequence", "band", "gene", "transcript", "cds", "exon", "repeat", "simple", "misc", "variation", "somatic_variation", "structural_variation", "somatic_structural_variation", "constrained", "regulatory", "motif", "mane" |
| "BioType" | None | functional classification of "gene" or "transcript" features to fetch; allowed value includes "protein_coding" |
| "DBType" | "core" | database type to retrieve features from; allowed values include: "core", "otherfeatures" |
| "SOTerm" | None | Sequence Ontology term to restrict the variants found |
| "TrimDownstream" | False | whether to return features which overlap the downstream end of the region |
| "TrimUpstream" | False | whether to return features which overlap the upstream end of the region |
| "VariantSet" | None | short name of a set to restrict the variants found such as "ClinVar" and "ph_uniprot"; list of short names are found here |
Retrieve a nucleotide sequence for a specified region of human chromosome 13:
| In[1]:= |
| Out[1]= |  |
Get a result as the BioSequence object:
| In[2]:= |
| Out[2]= |
Find genes that overlap a specified region of human chromosome 17:
| In[3]:= |
| Out[3]= |  |
Find genomic variations that overlap a specified region of human chromosome 6:
| In[4]:= |
| Out[4]= | 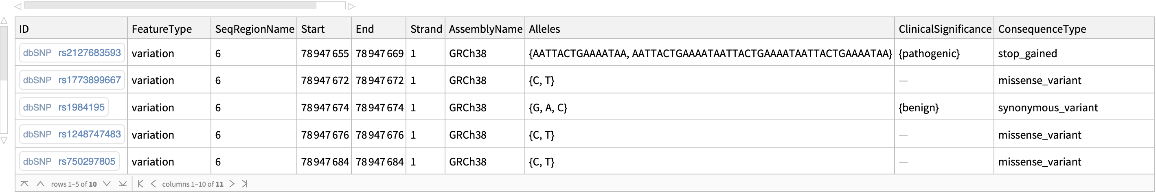 |
Use the NCBIGenomicSNPData resource function to retrieve more information on a selected variation:
| In[5]:= |
| Out[5]= |  |
Find its clinical significance:
| In[6]:= |
| Out[6]= |
Use the Species option to specify the organism of the genomics feature:
| In[7]:= |
| Out[7]= |  |
Use the Assembly option to specify the version of the genomics assembly:
| In[8]:= |
| Out[8]= |  |
Use the Mask option to retrieve the masked genome sequence where repeats are shown as lower cased characters:
| In[9]:= |
| Out[9]= |  |
Use the Strand option to retrieve the complementary DNA sequence:
| In[10]:= |
| Out[10]= |  |
Use the Feature option to selectively retrieve bands and transcripts associated with the given genomic region:
| In[11]:= |
| Out[11]= | 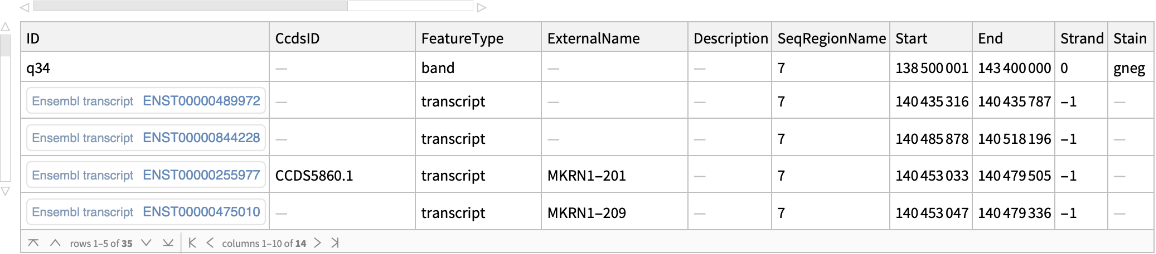 |
Use the BioType option to retrieve protein coding genes associated with the given genomic region:
| In[12]:= |
| Out[12]= | 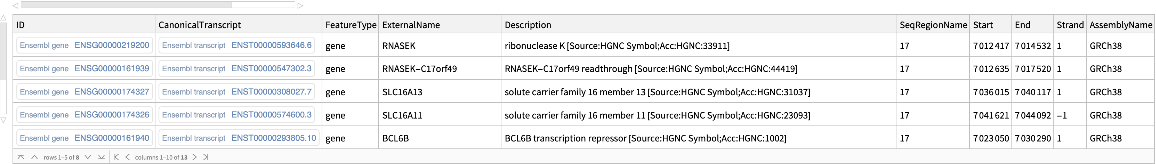 |
Use the DBType option to retrieve additional gene features associated with the given genomic region:
| In[13]:= |
| Out[13]= | 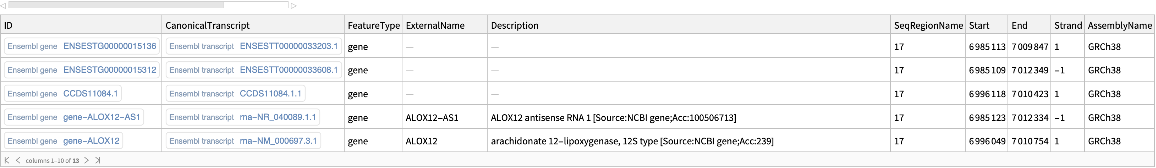 |
Use the SOTerm option to retrieve missense variants (SO:0001583) associated with the given genomic region:
| In[14]:= |
| Out[14]= | 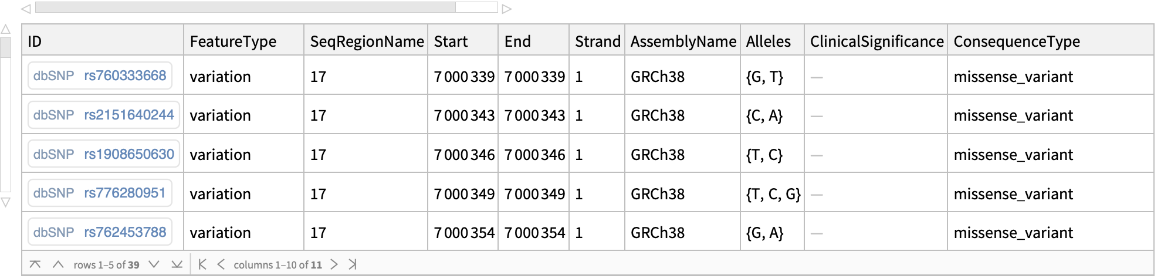 |
Use the TrimUpstream option to retrieve genes that overlap with the given genomic region, but not with the downstream region:
| In[15]:= |
| Out[15]= | 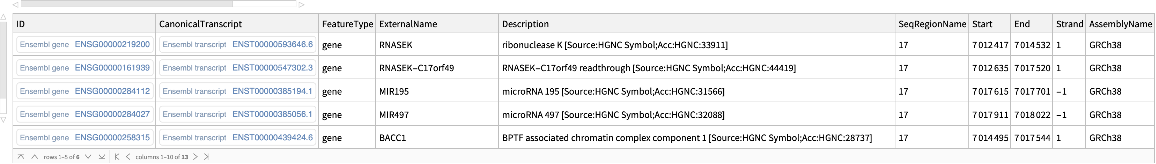 |
Use the TrimUpstream option to retrieve genes that overlap with the given genomic region, but not with the upstream region:
| In[16]:= |
| Out[16]= | 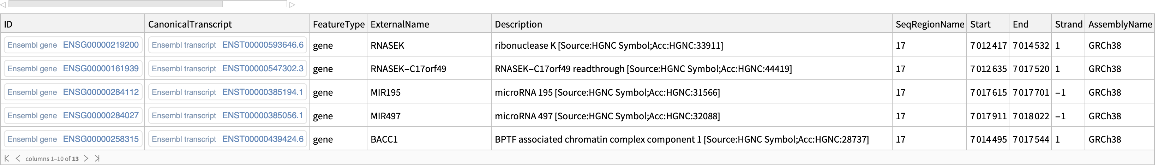 |
Use the VariantSet option to retrieve variants with ClinVar annotation associated with the given genomic region:
| In[17]:= |
| Out[17]= |  |
Find regulatory regions of the human chromosome 1:
| In[18]:= |
| Out[18]= | 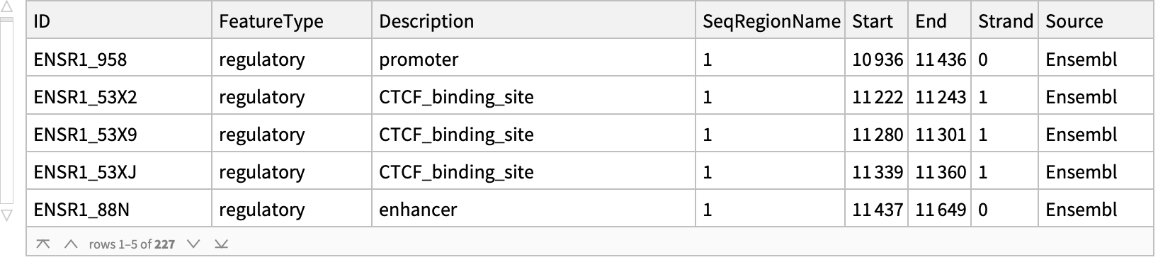 |
Group regions by the type of regulatory features:
| In[19]:= |
| Out[19]= | 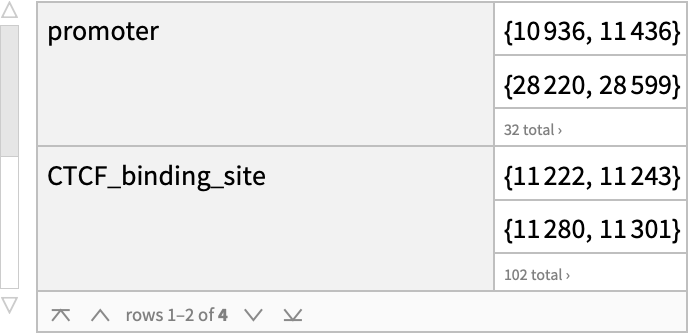 |
Visualize regulatory regions using the circular diagram illustrating their chromosome positions:
| In[20]:= | ![regulatoryregionbytypecol[col_, regdat_] := regdat[[1]] -> Riffle[Style[{Subtract[#[[2]], #[[1]]], 1}, GrayLevel[.2]] & /@ Transpose[{Prepend[regdat[[2]][[All, 2]], 0], Append[regdat[[2]][[All, 1]], 1000000]}], Style[{Subtract @@ Reverse[#], 1}, col] & /@ regdat[[2]]]](https://www.wolframcloud.com/obj/resourcesystem/images/cf2/cf2c0609-4db9-487a-ade6-528af443a10e/62ae9db91a09d259.png) |
| In[21]:= |
| In[22]:= | ![Show[SectorChart[regulatoryregioncolored[[1, 2]], SectorSpacing -> .1,
ChartBaseStyle -> EdgeForm[None], Epilog -> {Text[
Style[regulatoryregioncolored[[1, 1]], GrayLevel[.9], 10], {0, 4 - .2}], MapIndexed[
Text[Style[#1, GrayLevel[.9], 10], {0, #2[[1]] - .8/#2[[1]]}] &, regulatoryregioncolored[[2 ;; 4, 1]]]}, PolarAxes -> {True, False}, SectorOrigin -> {{Pi/2, Automatic}, 3.4},
PlotRange -> Full], SectorChart[regulatoryregioncolored[[2 ;; 4, 2]], SectorSpacing -> .1, ChartBaseStyle -> EdgeForm[None], PlotRange -> Full, SectorOrigin -> {{Pi/2, Automatic}, 0.1}]]](https://www.wolframcloud.com/obj/resourcesystem/images/cf2/cf2c0609-4db9-487a-ade6-528af443a10e/20eb1fd82b770d3d.png) |
| Out[22]= | 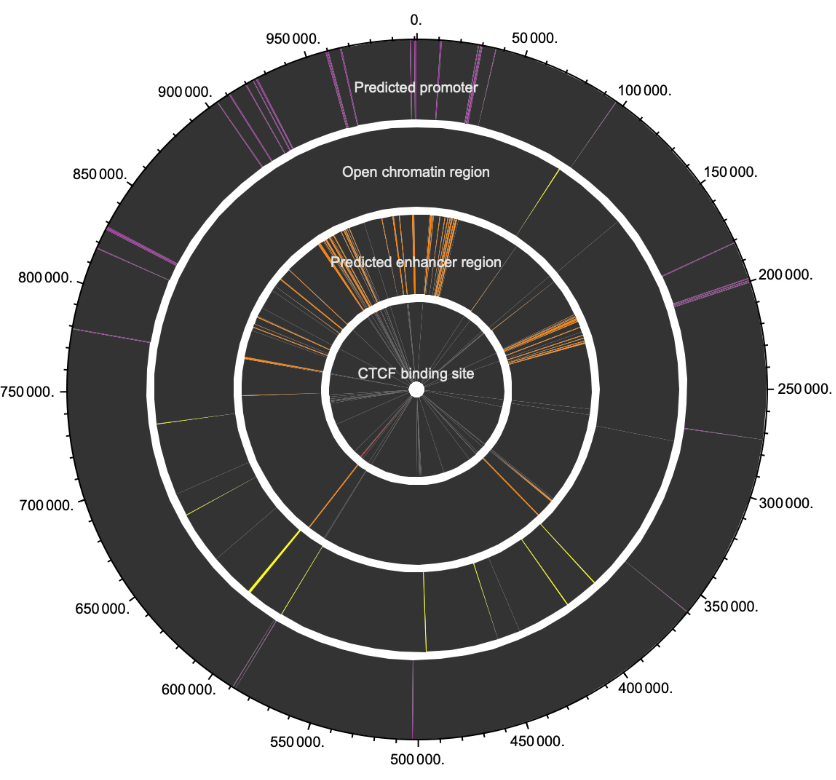 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License