Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Integrate a protein sequence into an existing phylogenetic tree of a protein family
ResourceFunction["PANTHERTreeGrafter"][seq, "Dataset"] gives the annotation result of the input seq in a dataset form. | |
ResourceFunction["PANTHERTreeGrafter"][seq, "Tabular"] gives the annotation result of the input seq in a tabular form. | |
ResourceFunction["PANTHERTreeGrafter"][seq, "TreeGraphic"] gives the protein family tree graphic highlighting the location of the grafted seq. | |
ResourceFunction["PANTHERTreeGrafter"][seq, "Tree"] gives the protein family tree highlighting the location of the grafted seq. | |
ResourceFunction["PANTHERTreeGrafter"]["Species"] gives the dataset of supported species. |
| "Species" | None | specified species to filter the result; accepted values include the scientific names, common names or NCBI Taxonomy ID of species found in the dataset retrieved by PANTHERTreeGrafter["Species"] |
Retrieve the annotation result for a protein sequence:
| In[1]:= | ![ResourceFunction[
"PANTHERTreeGrafter", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["MKVLWAALLVTFLAGCQAKVEQAVETEPEPELRQQTEWQSGQRWELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTPVAEETRARLSKELQAAQARLGADMEDVCGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVYQAGAREGAERGLSAIRERLGPLVEQGRVRAATVGSLAGQPLQERAQAWGERLRARMEEMGSRTRDRLDEVKEQVAEVRAKLEEQAQQIRLQAEAFQARLKSWFEPLVEDMQRQWAGLVEKVQAAVGTSAAPVPSDNH", "Dataset"]](https://www.wolframcloud.com/obj/resourcesystem/images/e89/e894987c-4c1b-4cc7-a107-bd12ff9d6c7c/4e439fa0a76ce91f.png) |
| Out[1]= |
Retrieve the annotation result in a tabular form instead:
| In[2]:= | ![ResourceFunction[
"PANTHERTreeGrafter", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["MKVLWAALLVTFLAGCQAKVEQAVETEPEPELRQQTEWQSGQRWELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTPVAEETRARLSKELQAAQARLGADMEDVCGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVYQAGAREGAERGLSAIRERLGPLVEQGRVRAATVGSLAGQPLQERAQAWGERLRARMEEMGSRTRDRLDEVKEQVAEVRAKLEEQAQQIRLQAEAFQARLKSWFEPLVEDMQRQWAGLVEKVQAAVGTSAAPVPSDNH", "Tabular"]](https://www.wolframcloud.com/obj/resourcesystem/images/e89/e894987c-4c1b-4cc7-a107-bd12ff9d6c7c/281f803694de895c.png) |
| Out[2]= | 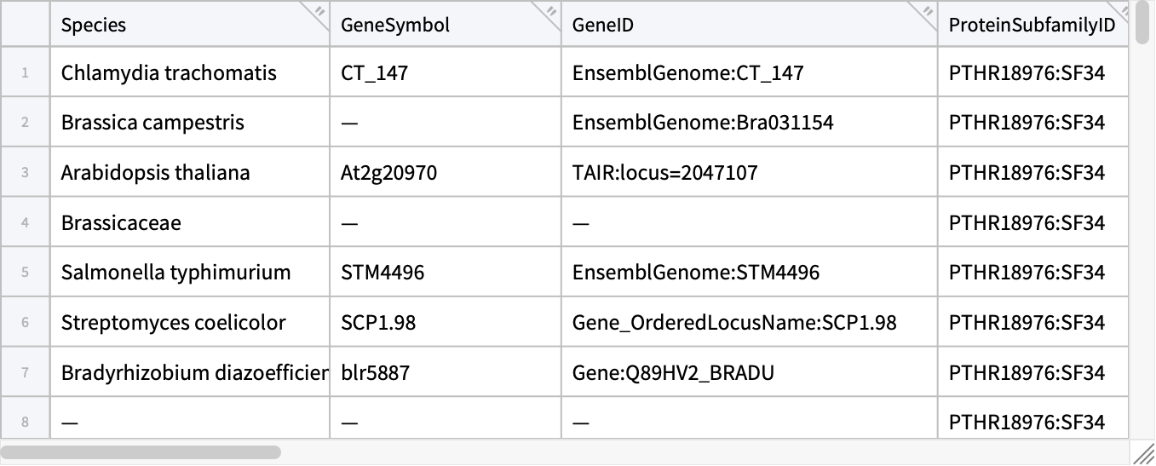 |
Highlight the location of the grafted sequence on the protein family phylogenetic tree graphic. Each node is labeled with the corresponding accession from the dataset. Hover over or click on any nodes to find associated species information:
| In[3]:= | ![ResourceFunction[
"PANTHERTreeGrafter", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["MKVLWAALLVTFLAGCQAKVEQAVETEPEPELRQQTEWQSGQRWELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTPVAEETRARLSKELQAAQARLGADMEDVCGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVYQAGAREGAERGLSAIRERLGPLVEQGRVRAATVGSLAGQPLQERAQAWGERLRARMEEMGSRTRDRLDEVKEQVAEVRAKLEEQAQQIRLQAEAFQARLKSWFEPLVEDMQRQWAGLVEKVQAAVGTSAAPVPSDNH", "TreeGraphic"]](https://www.wolframcloud.com/obj/resourcesystem/images/e89/e894987c-4c1b-4cc7-a107-bd12ff9d6c7c/3e20915e04b5e8ad.png) |
| Out[3]= |  |
Retrieve the protein family tree, highlighting the location of the grafted sequence. Hover over or click on any tree elements to find associated species information:
| In[4]:= | ![ResourceFunction[
"PANTHERTreeGrafter", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["MKVLWAALLVTFLAGCQAKVEQAVETEPEPELRQQTEWQSGQRWELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTPVAEETRARLSKELQAAQARLGADMEDVCGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVYQAGAREGAERGLSAIRERLGPLVEQGRVRAATVGSLAGQPLQERAQAWGERLRARMEEMGSRTRDRLDEVKEQVAEVRAKLEEQAQQIRLQAEAFQARLKSWFEPLVEDMQRQWAGLVEKVQAAVGTSAAPVPSDNH", "Tree"]](https://www.wolframcloud.com/obj/resourcesystem/images/e89/e894987c-4c1b-4cc7-a107-bd12ff9d6c7c/28a47e673b5751e9.png) |
| Out[4]= | 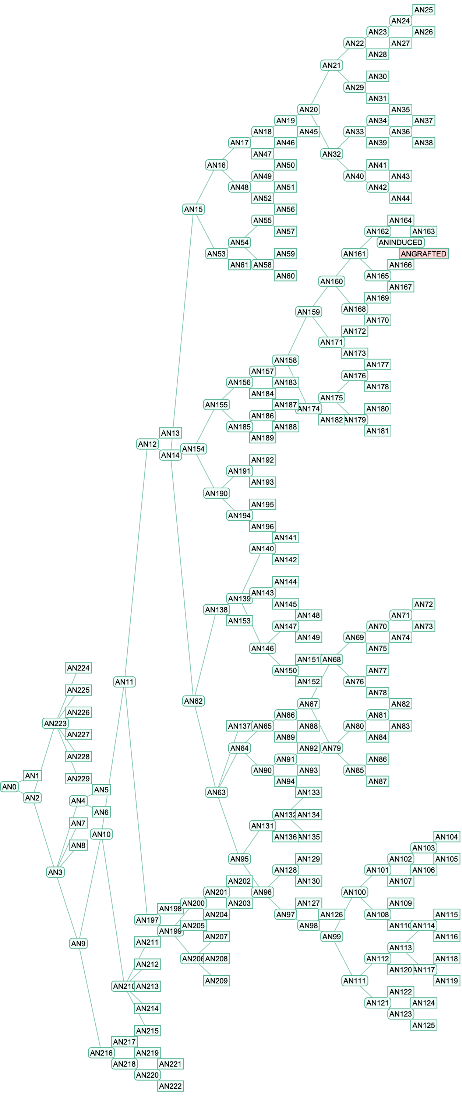 |
Retrieve the annotation result for a protein sequence:
| In[5]:= | ![pantherdataset = ResourceFunction[
"PANTHERTreeGrafter", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"][BioSequence[
"Peptide", "ILQHNQNMSGLEKVSKISPCDVSLETSDICKCSIGKLHKSVSSANTCGIFSTASGKSVQVSDASLQNARQVFSEIEDSTKQVFSKVLFKSNEHSD", {}], "Dataset"]](https://www.wolframcloud.com/obj/resourcesystem/images/e89/e894987c-4c1b-4cc7-a107-bd12ff9d6c7c/65218103a5d2c81a.png) |
| Out[5]= |
Explore annotated terms associated with the grafted sequence:
| In[6]:= |
| Out[6]= | 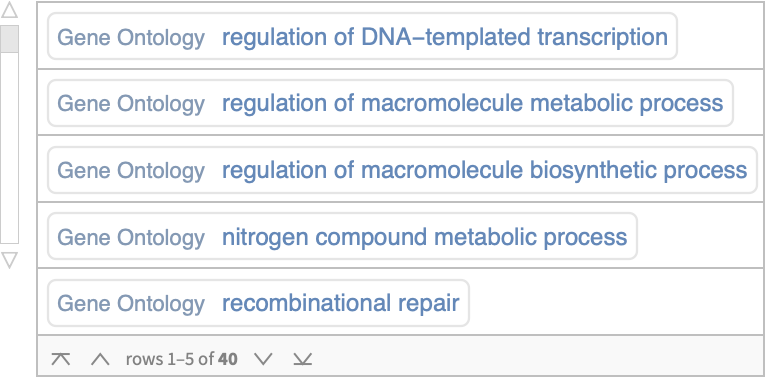 |
Find the associated human gene:
| In[7]:= |
| Out[7]= |
Use the BioDBnetGeneData resource function to get more information on the gene:
| In[8]:= |
| Out[8]= | 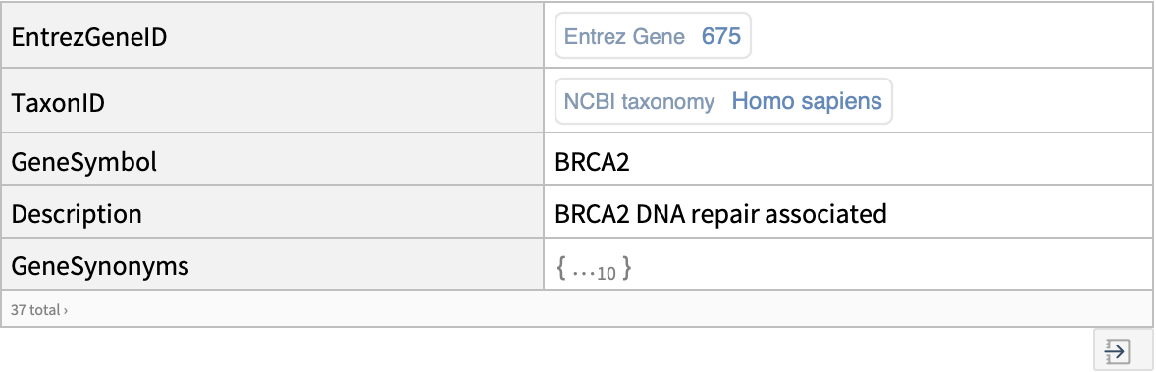 |
Retrieve the result of the tree analysis filtered for the Mus musculus species:
| In[9]:= | ![ResourceFunction[
"PANTHERTreeGrafter", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["MKVLWAALLVTFLAGCQAKVEQAVETEPEPELRQQTEWQSGQRWELALGRFWDYLRWVQTLSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTPVAEETRARLSKELQAAQARLGADMEDVCGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVYQAGAREGAERGLSAIRERLGPLVEQGRVRAATVGSLAGQPLQERAQAWGERLRARMEEMGSRTRDRLDEVKEQVAEVRAKLEEQAQQIRLQAEAFQARLKSWFEPLVEDMQRQWAGLVEKVQAAVGTSAAPVPSDNH", "Tree", "Species" -> "Mus musculus"]](https://www.wolframcloud.com/obj/resourcesystem/images/e89/e894987c-4c1b-4cc7-a107-bd12ff9d6c7c/41f76a5a150b91bb.png) |
| Out[9]= | 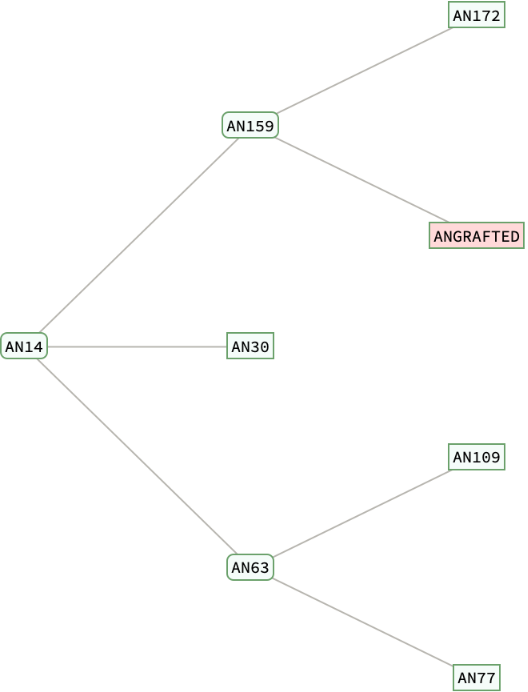 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License