Details and Options
ResourceFunction["FCGRImage"][str] uses a default pixelation level of 7 and works with the square having "A" as the lower-right vertex, proceeding counterclockwise with "T", "G" and "C".
ResourceFunction["FCGRImage"][str,k] gives an image of dimension 2k×2k.
For ResourceFunction["FCGRImage"][str,k,bases], the third argument must be a permutation of the list {"A","C","G","T"}.
Any occurrence of the character "U" in str is converted to "T".
All characters in str are converted to uppercase.
All characters in the converted input str other than {"A","C","G","T"} will be removed.
There is an "off-centered" asymmetry of the nucleotide positions from using grids that have side lengths of powers of 2. This will be illustrated in examples. It can be removed by enlarging the grids by one unit in each dimension.
By default, the asymmetry is not removed. The body of literature on the FCGR gives no indication of doing this. Perhaps more importantly, empirical evidence suggests that results are better when working with the slightly off-centered version.
ResourceFunction["FCGRImage"][bioseq] gives the FCGR image of a
BioSequence expression that is of type
"DNA",
"RNA",
"CircularDNA" or
"CircularRNA". The
BioSequence expression should not have any degenerate letters.

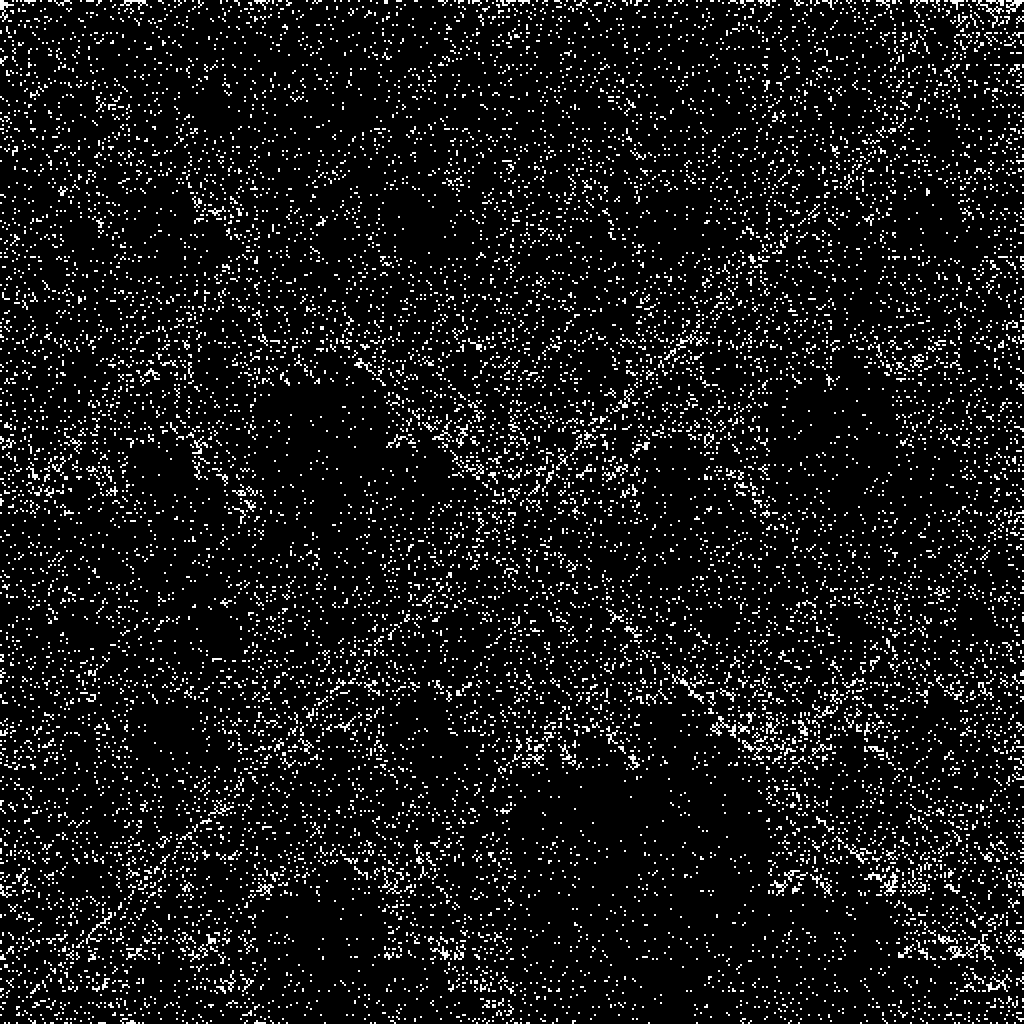
![SeedRandom[12345678];
charstring = ToString[RandomChoice[{"A", "T", "C", "G"}, 2000]];
ResourceFunction["FCGRImage"][charstring, ImageSize -> 200]](https://www.wolframcloud.com/obj/resourcesystem/images/6d8/6d81e10b-9ab8-4599-bd6f-1c63fbaf8db0/0f559c9ae8b6a6f4.png)




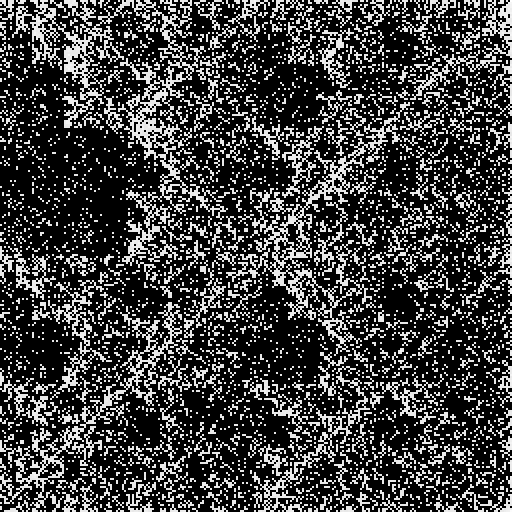

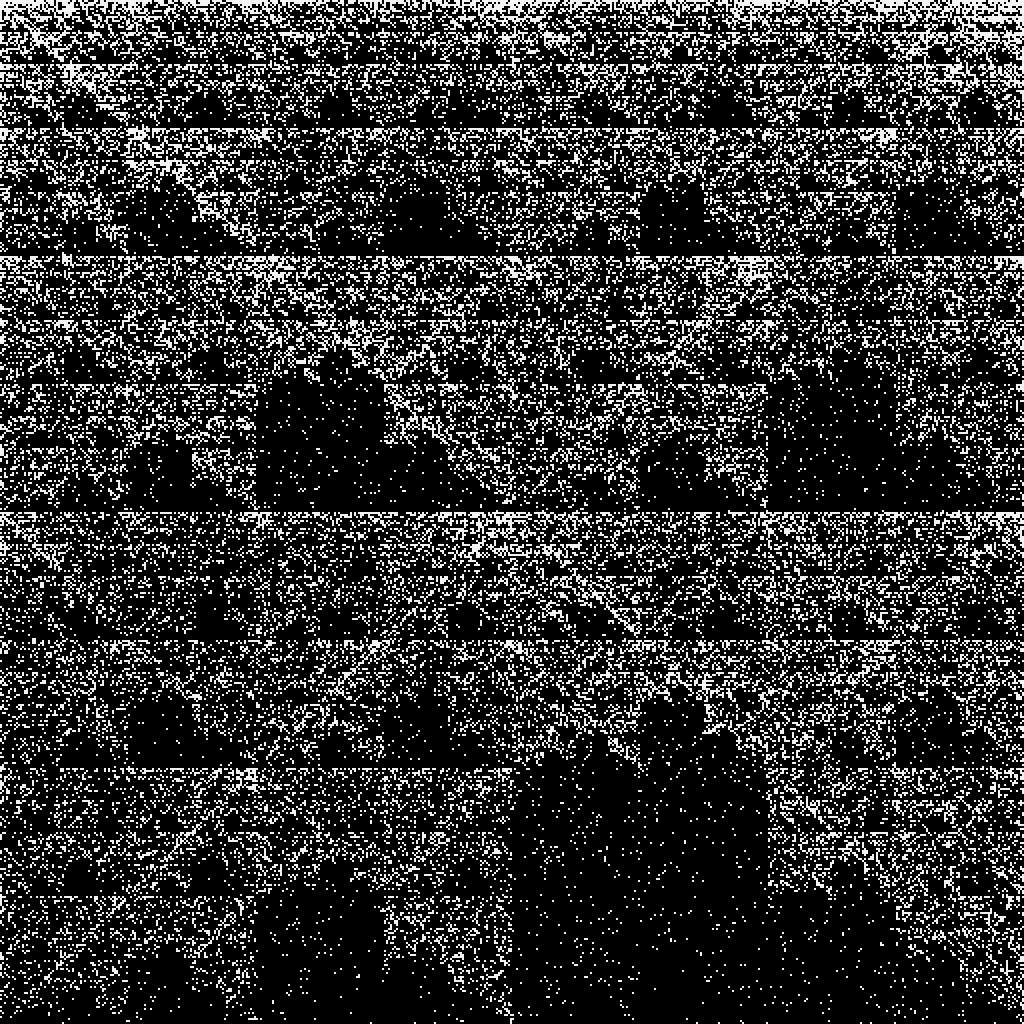
![gene = First[
URLExecute[
"https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi", <|
"db" -> "nuccore", "id" -> "U01317.1", "rettype" -> "fasta", "retmode" -> "text"|>, "FASTA"]];](https://www.wolframcloud.com/obj/resourcesystem/images/6d8/6d81e10b-9ab8-4599-bd6f-1c63fbaf8db0/5aaef3fe2a2d3414.png)
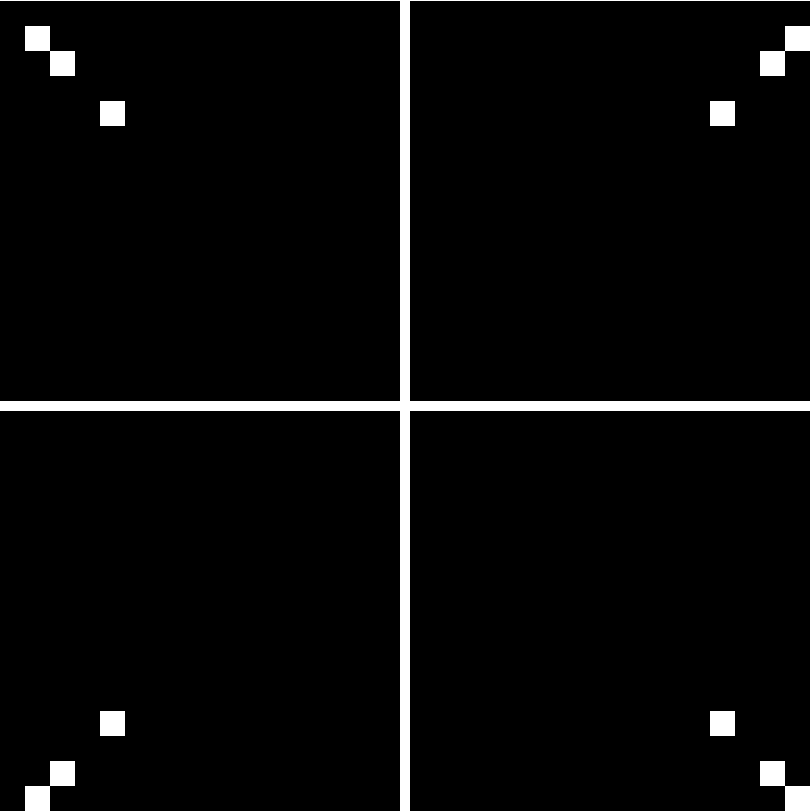
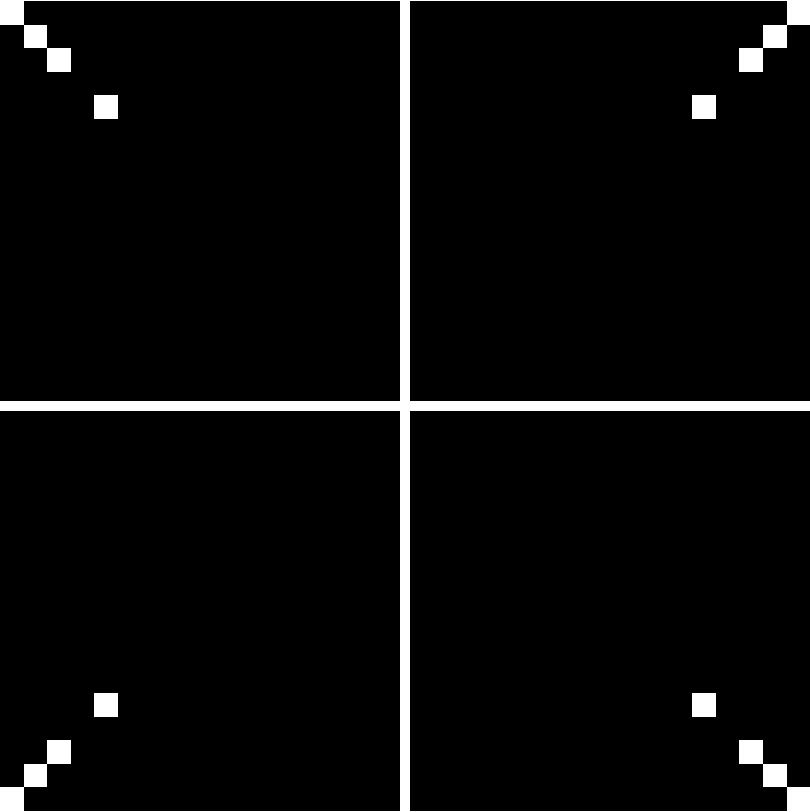
![Grid[Partition[
Map[ResourceFunction["FCGRImage"][#, 4, ImageSize -> 200] &, {"AAAA", "TTTT", "CCCC", ",GGGG"}], 2]]](https://www.wolframcloud.com/obj/resourcesystem/images/6d8/6d81e10b-9ab8-4599-bd6f-1c63fbaf8db0/73587e6bb3058d45.png)
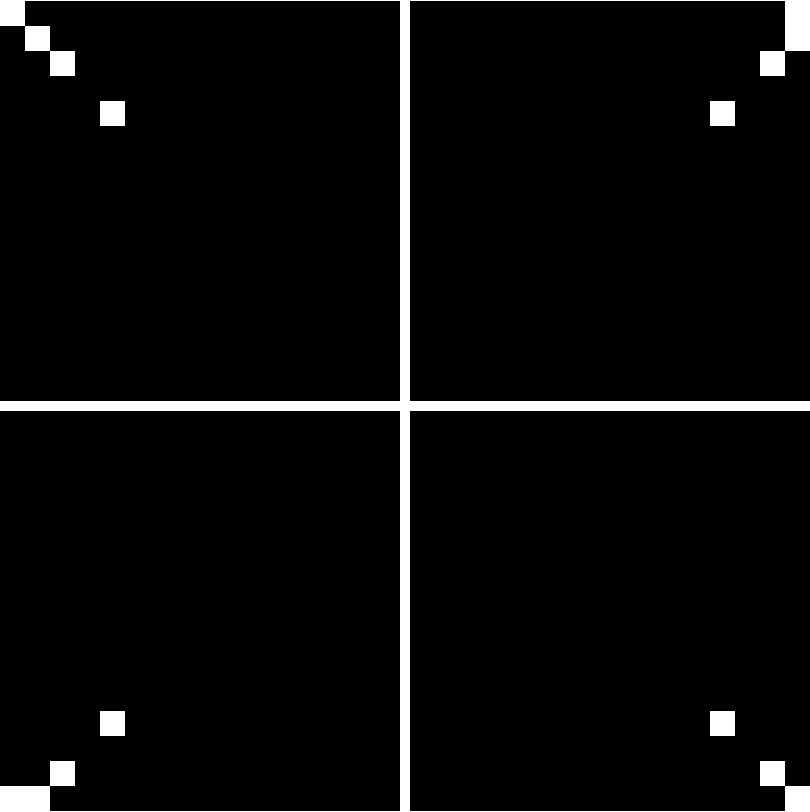
![Grid[Partition[
Map[ResourceFunction["FCGRImage"][#, 4, "Centered" -> True, ImageSize -> 200] &, {"AAAA", "TTTT", "CCCC", ",GGGG"}], 2]]](https://www.wolframcloud.com/obj/resourcesystem/images/6d8/6d81e10b-9ab8-4599-bd6f-1c63fbaf8db0/3ef6118c58ea9b85.png)