Details and Options
EnsemblPhenotype is based on
Ensembl, which provides genomics information including phenotype annotations. Phenotypes are traits that are associated with the genomic variants.
To retrieve "PhenotypeAnnotations", specify the chromosome argument by a chromosome name such as "chromosome 1" or simply just "1" and the start and end positions of the genome sequence by the integers.
The following option can be given:
| "Species" | "human" | species for which to query; should be one of selected TaxonomicSpecies entities or names of species |
Supported species include: human (Homo sapiens), cat (Felis catus), chicken (Gallus gallus), cow (Bos taurus), dog (Canis lupus familiaris), goat (Capra hircus), horse (Equus caballus), mouse (Mus musculus), pig (Sus scrofa), rat (Rattus norvegicus), sheep (Ovis aries) and zebrafish (Danio rerio).
ResourceFunction["EnsemblPhenotype"][phenotype] is equivalent to ResourceFunction["EnsemblPhenotype"][phenotype,"GenomicFeatures"].
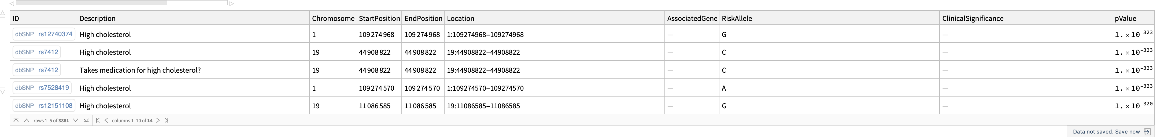

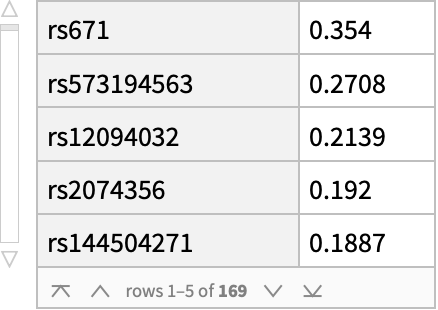
![coffeConsumptionRateAffectingSNP = snps[Select[! MissingQ[#BetaCoefficient] && ! StringFreeQ[#BetaCoefficient, "unit" | "cups"] &]][
All, {#ID[
"ExternalID"], (StringTrim[
StringSplit[#BetaCoefficient, i : "increase" | "decrease" :> i], " unit" | " NR" | " cups" ~~ __] /. {{d_, "increase"} :> ToExpression@d, {d_, "decrease"} :> -ToExpression@d})} &][
GroupBy[First -> Last], MeanAround][
ReverseSortBy[# /. a_Around :> a[[1]] &]]](https://www.wolframcloud.com/obj/resourcesystem/images/69f/69febbba-e5f6-4cd2-964b-7462e3ad4af8/69a1a7fcd5791544.png)
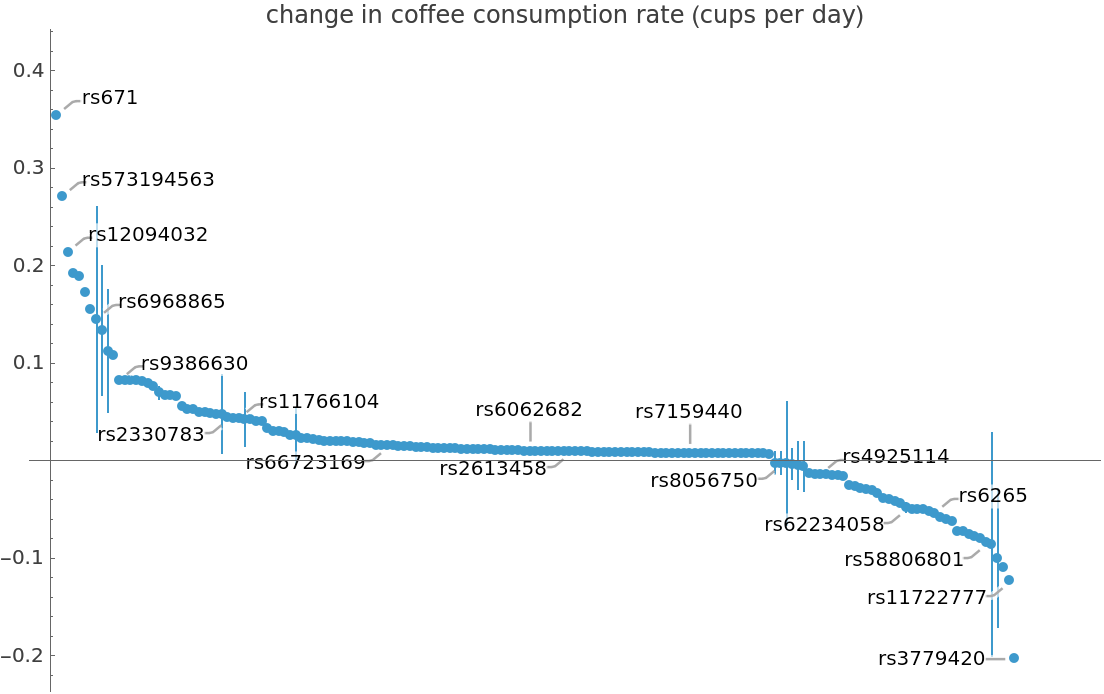
![ListPlot[coffeConsumptionRateAffectingSNP, PlotRange -> Full, Ticks -> {None, Automatic}, PlotLabel -> "change in coffee consumption rate (cups per day)"]](https://www.wolframcloud.com/obj/resourcesystem/images/69f/69febbba-e5f6-4cd2-964b-7462e3ad4af8/5bb830694b018b22.png)
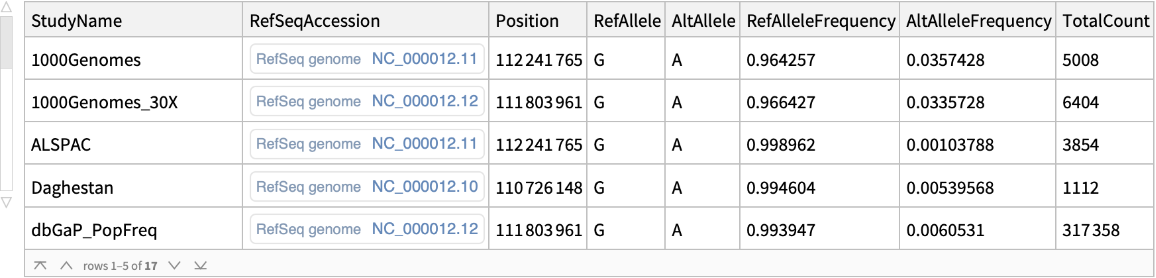
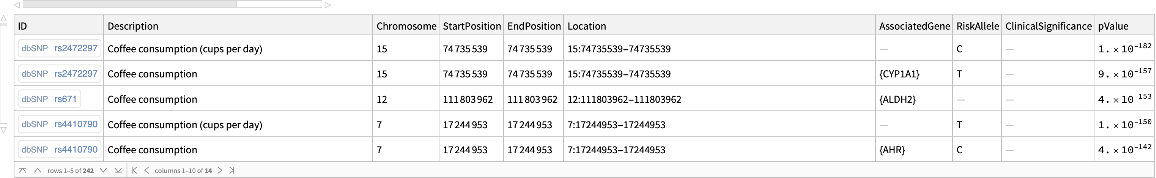
![coffeConsumptionRateAffectingGene = Dataset[GroupBy[
Flatten@Normal@
snps[Select[! MissingQ[#AssociatedGene] && ! MissingQ[#BetaCoefficient] && ! StringFreeQ[#BetaCoefficient, "unit" | "cups"] &]][All, Thread[(StringTrim[
StringSplit[#BetaCoefficient, i : "increase" | "decrease" :> i], " unit" | " NR" | " cups" ~~ __] /. {{d_, "increase"} :> ToExpression@d, {d_, "decrease"} :> -ToExpression@d}) -> #AssociatedGene] &][
ReverseSortBy[Mean[#] &]], Last -> First, MeanAround]]](https://www.wolframcloud.com/obj/resourcesystem/images/69f/69febbba-e5f6-4cd2-964b-7462e3ad4af8/390c85da1f509ff5.png)

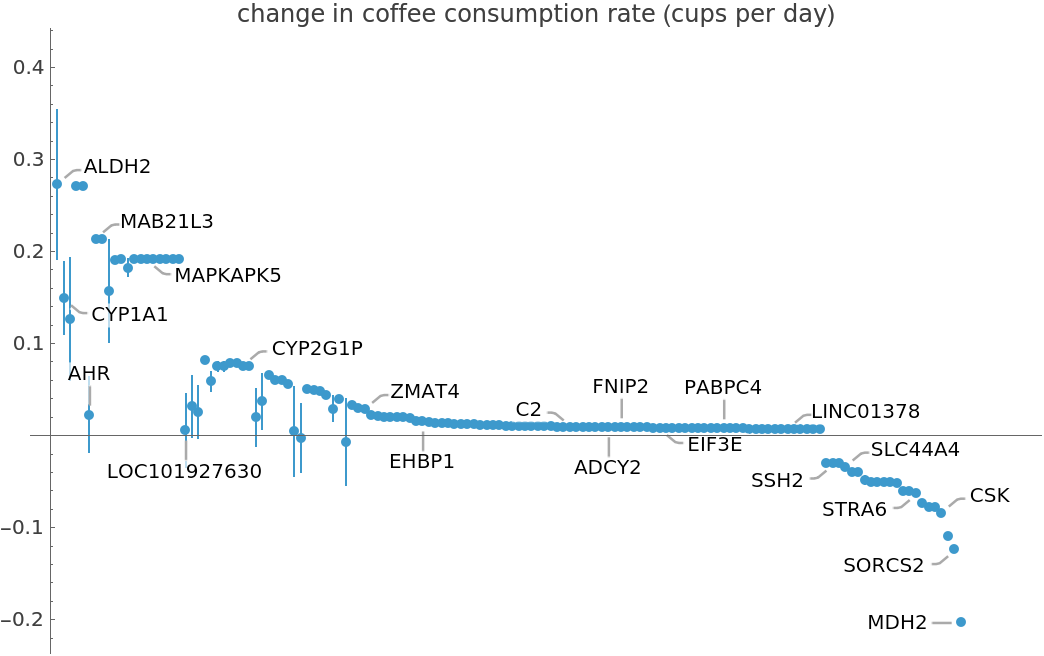
![ListPlot[coffeConsumptionRateAffectingGene, PlotRange -> Full, Ticks -> {None, Automatic}, PlotLabel -> "change in coffee consumption rate (cups per day)"]](https://www.wolframcloud.com/obj/resourcesystem/images/69f/69febbba-e5f6-4cd2-964b-7462e3ad4af8/00004e08a89e336d.png)
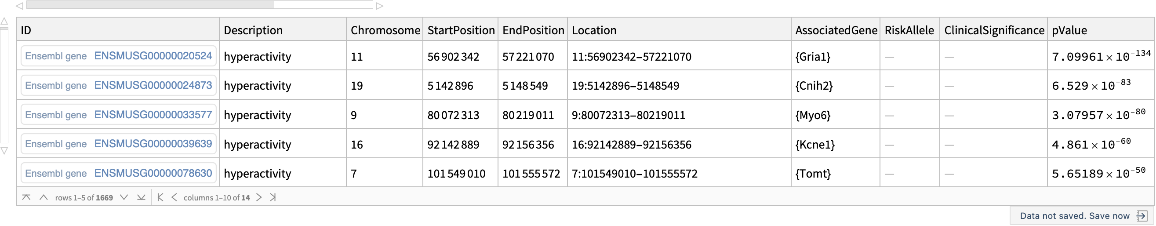
![ResourceFunction[
"EnsemblPhenotype", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["hyperactivity", "GenomicFeatures", "Species" -> Entity["TaxonomicSpecies", "MusMusculus::y84t7"]]](https://www.wolframcloud.com/obj/resourcesystem/images/69f/69febbba-e5f6-4cd2-964b-7462e3ad4af8/2a7ef69fb6bdcff8.png)