Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Look up Ensembl identifiers or external symbols and provide cross references in other databases
ResourceFunction["EnsemblCrossReference"][id] looks up id and retrieves its cross references. | |
ResourceFunction["EnsemblCrossReference"]["DatabasesBySpecies"] retrieves the list of supported databases by species. |
| "Species" | "human" | species for which to query; should be one of selected TaxonomicSpecies entities or names of species; use EnsemblCrossReference["DatabasesBySpecies"] to get the list supported species |
| "Database" | All | specifies from which to retrieve references; use EnsemblCrossReference["DatabasesBySpecies"] to get the list supported databases |
Retrieve cross references for an Ensembl gene:
| In[1]:= |
| Out[1]= |  |
Retrieve cross references for a specified gene symbol:
| In[2]:= |
| Out[2]= | 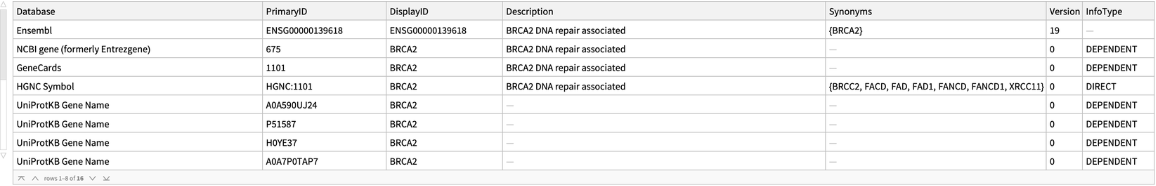 |
Find the list of supported external databases by species:
| In[3]:= |
| Out[3]= | 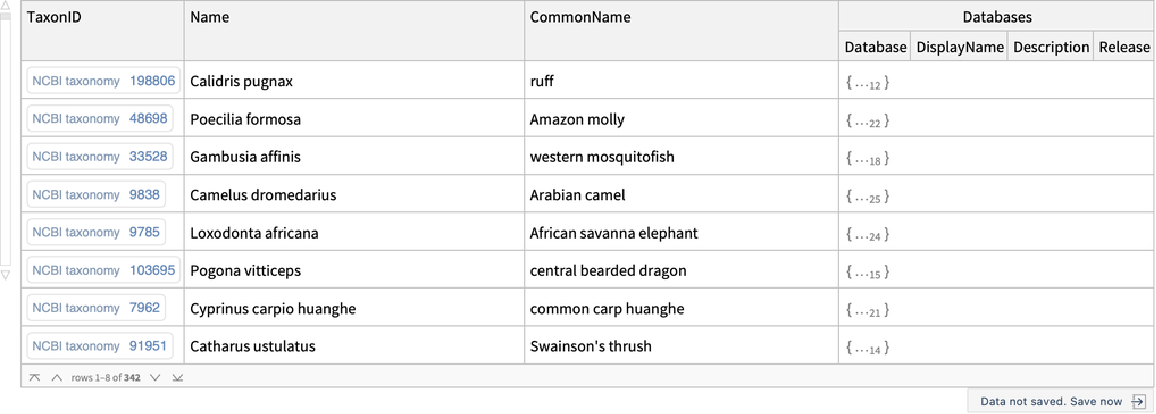 |
For ambiguous identifiers, EnsemblCrossReference looks up all potential references that match the identifier:
| In[4]:= |
| Out[4]= | 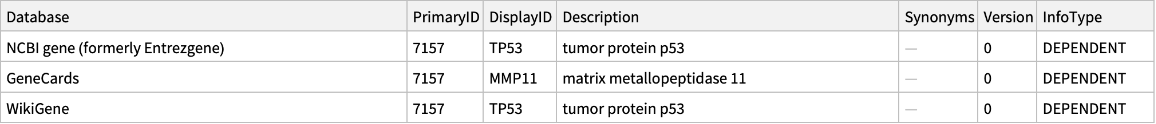 |
Use the "Species" option to specify the organism to query about:
| In[5]:= |
| Out[5]= | 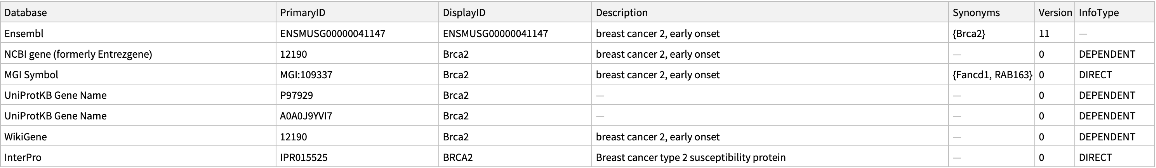 |
Use the "Database" option to specify the database from which to retrieve the cross reference:
| In[6]:= |
| Out[6]= | 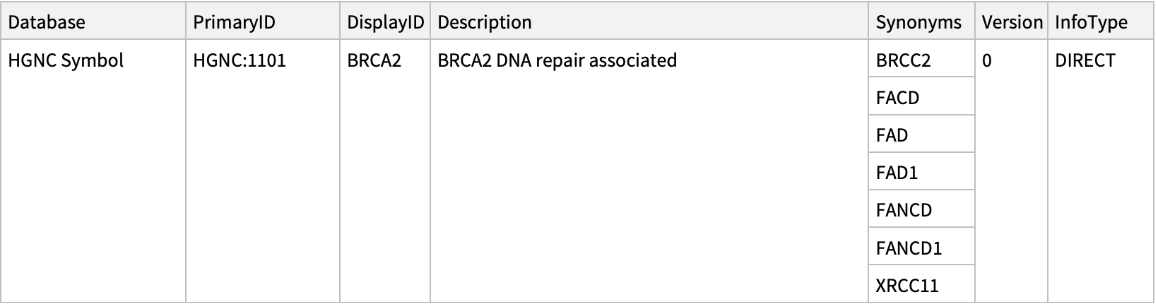 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License