Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Convert a given strand of DNA to a list of amino acids
ResourceFunction["DNAtoAminoAcid"][dna] converts the input DNA string to a list of amino acid entities. |
Convert the DNA strand “TACTTTTCGTCCGGTATAATT" to a list of amino acids:
| In[1]:= |
| Out[1]= |
Convert an invalid DNA strand with letters other than A/C/T/G:
| In[2]:= |
| Out[2]= |
Convert a DNA strand without a "TAC" present:
| In[3]:= |
| Out[3]= |
Create a visualization of the chain of amino acids by combining this with BioSequenceMoleculePlot:
| In[4]:= | ![ResourceFunction["BioSequenceMoleculePlot"][
BioSequence["Peptide", ResourceFunction["DNAtoAminoAcid"][
"GTATACTGGTCATAGCATTGACTGGTCCATGTACTTACCGCT"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/679/67954e72-53c2-4527-a7c0-68dd2ba1497e/1-0-0/2d14a69139f78b69.png) |
| Out[4]= | 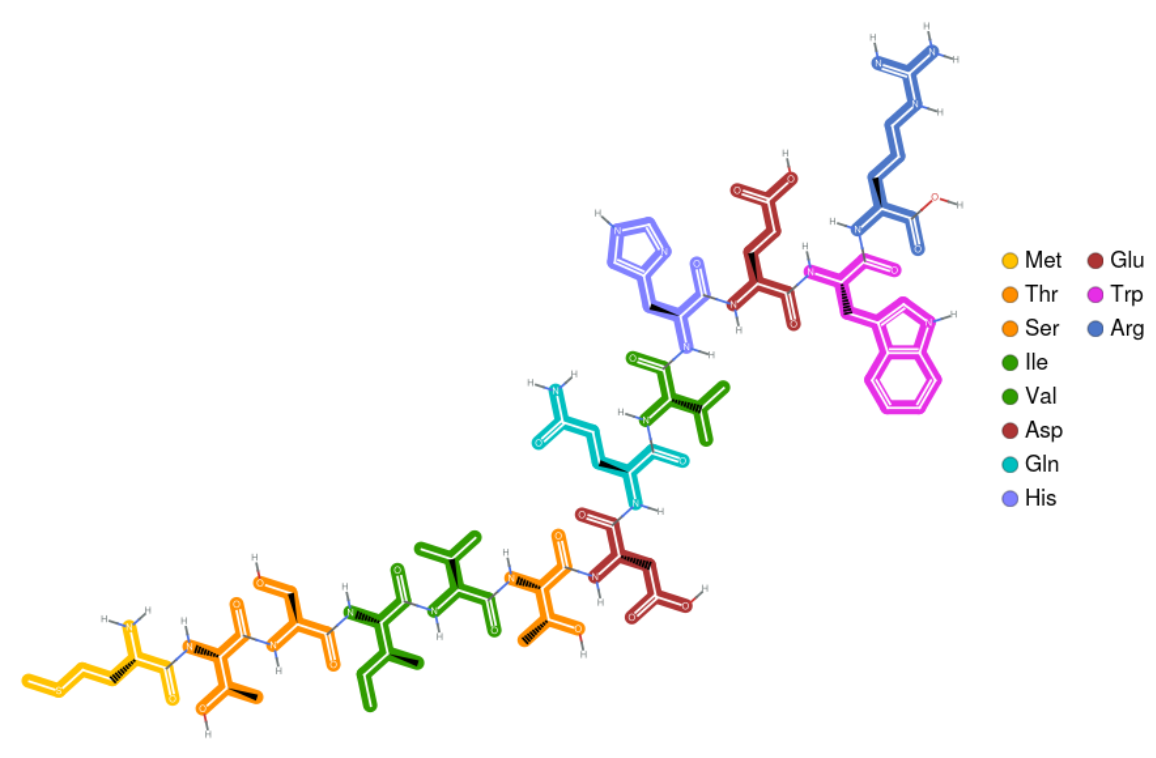 |
This work is licensed under a Creative Commons Attribution 4.0 International License