Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Visualize a biomolecular sequence in 3D with highlighted residues
ResourceFunction["BioSequenceMoleculePlot3D"][bioseq] creates a three-dimensional model of the biomolecular sequence bioseq, where all residues are highlighted. |
| "Highlighting" | Automatic | how to highlight the given BioSequence |
Visualize the structure of the "beefy meaty peptide":
| In[1]:= |
| Out[1]= | 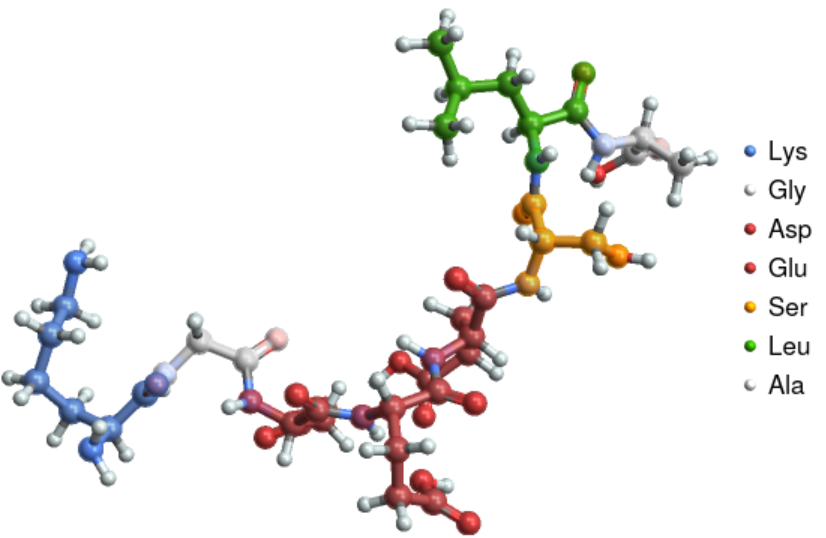 |
Visualize the structure of the circular peptide evolidine:
| In[2]:= |
| Out[2]= | 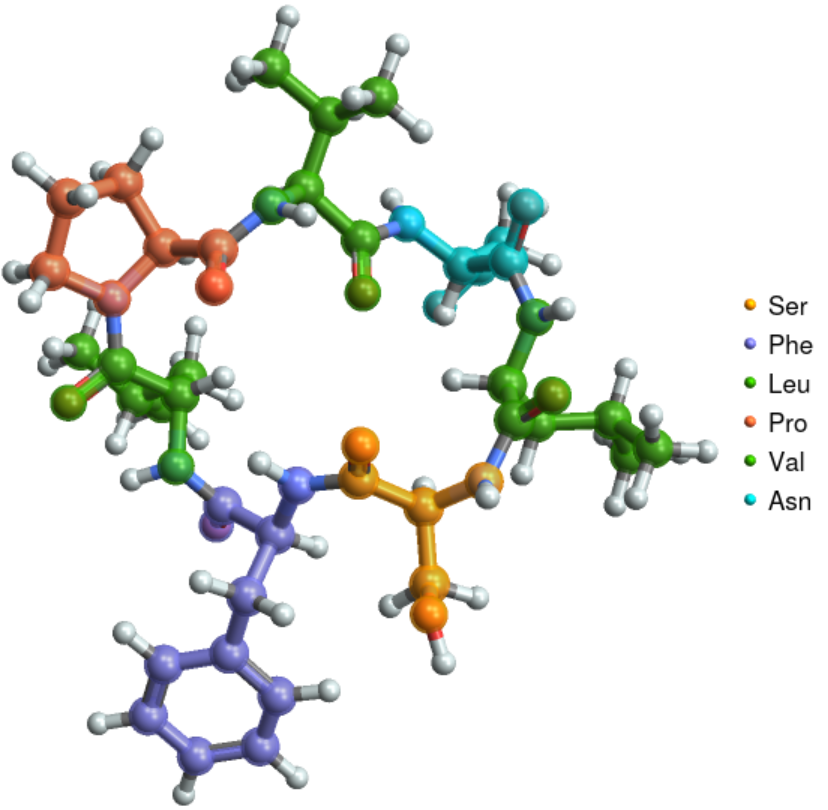 |
Compare an RNA and a circular RNA sequence:
| In[3]:= | ![{ResourceFunction["BioSequenceMoleculePlot3D"][
BioSequence["RNA", "AUCG"]], ResourceFunction["BioSequenceMoleculePlot3D"][
BioSequence["CircularRNA", "AUCG"]]} // GraphicsRow](https://www.wolframcloud.com/obj/resourcesystem/images/41a/41ac3ae7-dfed-4aa5-ad39-810d7dc9c3a8/4efca08380b8fd40.png) |
| Out[3]= | 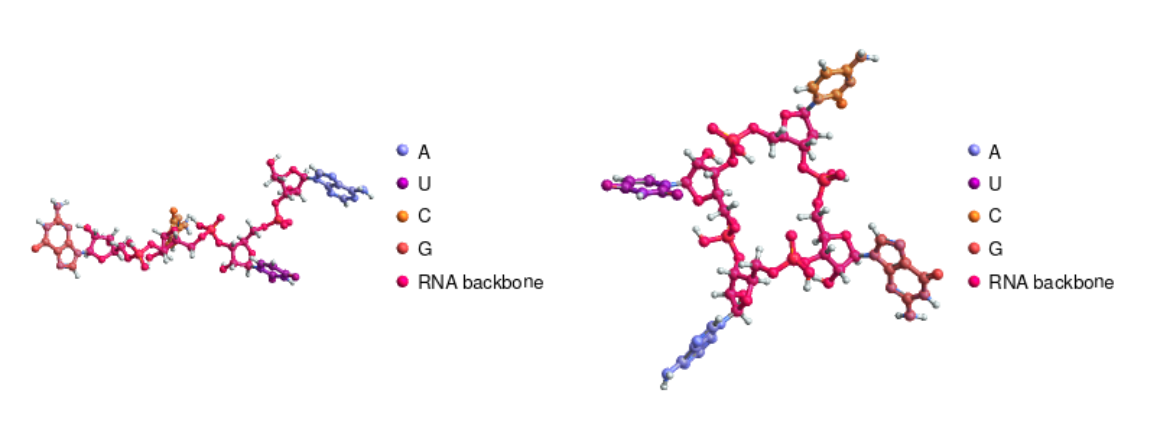 |
Highlight the sugars and phosphate groups together in a DNA-RNA hybrid strand:
| In[4]:= | ![ResourceFunction["BioSequenceMoleculePlot3D"][
BioSequence[
"HybridStrand", {BioSequence["DNA", "TGC"], BioSequence["RNA", "GUA"]}], "Highlighting" -> "ResidueBackbone"]](https://www.wolframcloud.com/obj/resourcesystem/images/41a/41ac3ae7-dfed-4aa5-ad39-810d7dc9c3a8/37aded448ff3ec4f.png) |
| Out[4]= | 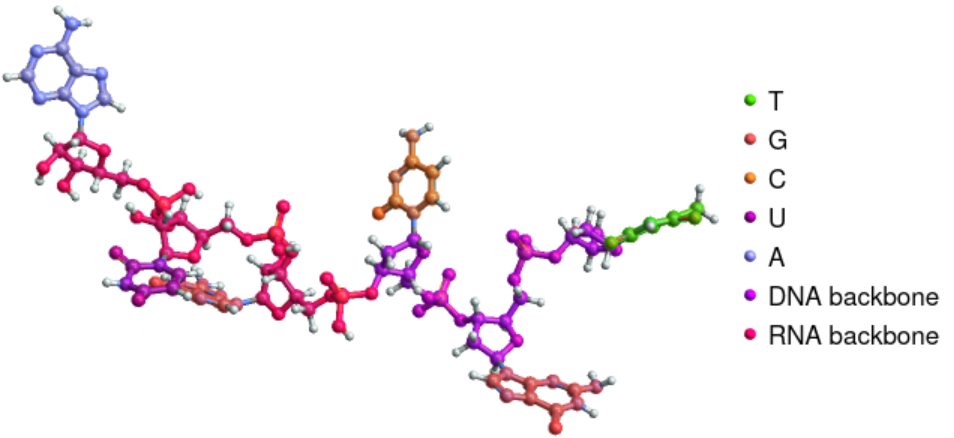 |
Highlight the sugars and phosphate groups separately:
| In[5]:= | ![ResourceFunction["BioSequenceMoleculePlot3D"][
BioSequence[
"HybridStrand", {BioSequence["DNA", "TGC"], BioSequence["RNA", "GUA"]}], "Highlighting" -> "ResidueSugar"]](https://www.wolframcloud.com/obj/resourcesystem/images/41a/41ac3ae7-dfed-4aa5-ad39-810d7dc9c3a8/4a4fa253429f5d5a.png) |
| Out[5]= | 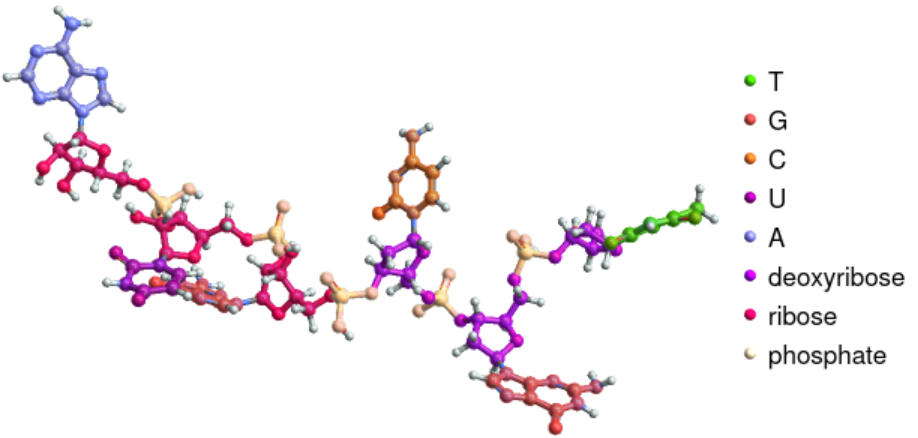 |
Highlight the amino acid residues in robustoxin, a component of the venom of the spider Atrax robustus:
| In[6]:= | ![ResourceFunction["BioSequenceMoleculePlot3D"][
BioSequence["CAKKRNWCGKNEDCCCPMKCIYAWYNQQGSCQTTITGLFKKC", Bond /@ {{1, 15}, {8, 20}, {14, 31}, {16, 42}}]]](https://www.wolframcloud.com/obj/resourcesystem/images/41a/41ac3ae7-dfed-4aa5-ad39-810d7dc9c3a8/3d0fa3d15f37190a.png) |
| Out[6]= | 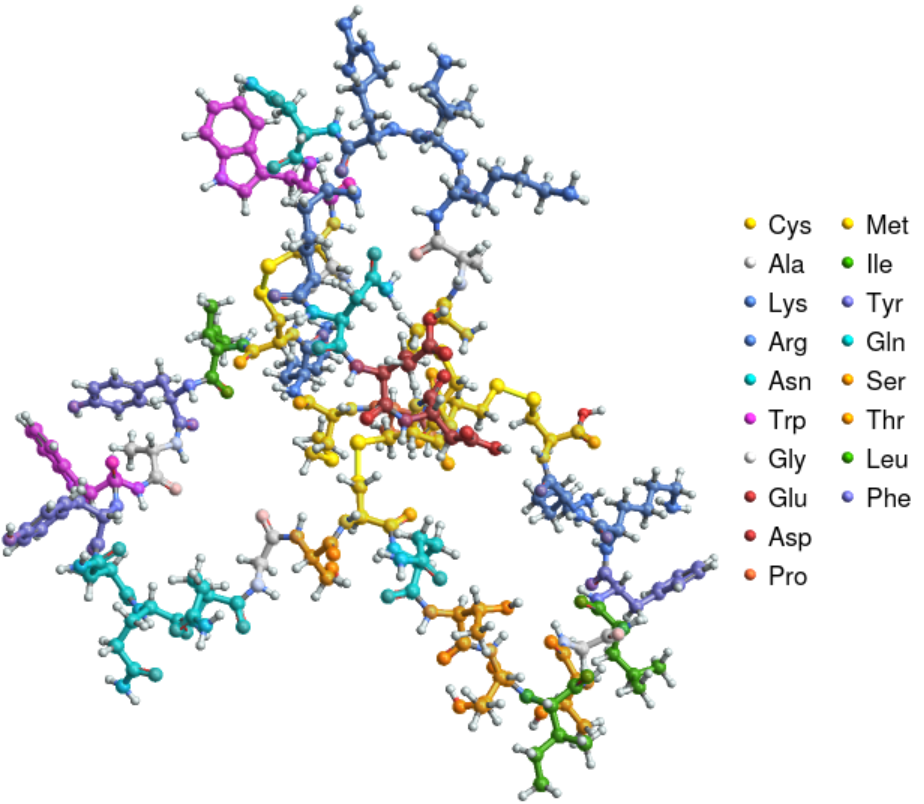 |
This work is licensed under a Creative Commons Attribution 4.0 International License