Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Generate the hydropathy plot of a peptide sequence
ResourceFunction["PeptideHydropathyPlot"][expr] gives the hydropathy plot of the peptide represented by expr. |
| WindowSize | Automatic | size of the window used for the moving average computation |
Hydropathy plot of a "Peptide" BioSequence:
| In[2]:= |
| Out[2]= | 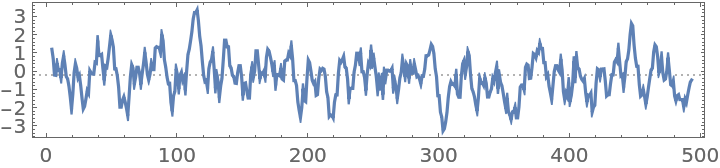 |
Hydropathy plot for bovine chymotrypsinogen A:
| In[3]:= | ![ResourceFunction[
"PeptideHydropathyPlot"]["CGVPAIQPVLSGLSRIVNGEEAVPGSWPWQVSLQDKTGFHFCGGSLINENWVVTAAHCGVTTSDVVVAGEFDQGSSSEKIQKLKIAKVFKNSKYNSLTINNDITLLKLSTAASFSQTVSAVCLPSASDDFAAGTTCVTTGWGLTRYTNANTPDRLQQASLPLLSNTNCKKYWGTKIKDAMICAGASGVSSCMGDSGGPLVCKKNGAWTLVGIVSWGSSTCSTSTPGVYARVTALVNWVQQTLAAN"]](https://www.wolframcloud.com/obj/resourcesystem/images/3a1/3a1da606-52f5-4b25-8b8c-e575fe48c1ca/02715bd7786016eb.png) |
| Out[3]= | 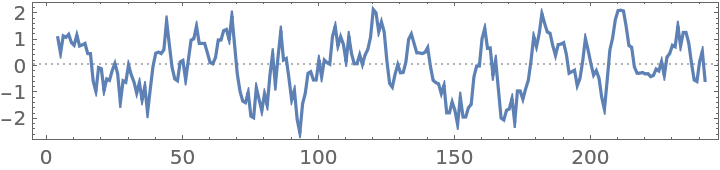 |
Use different window sizes for the hydropathy plot of bacteriorhodopsin:
| In[4]:= | ![Table[ResourceFunction["PeptideHydropathyPlot"][BioSequence[
"Peptide", "EAQITGRPEWIWLALGTALMGLGTLYFLVKGMGVSDPDAKKFYAITTLVPAIAFTMYLSMLLGYGLTMVPFGGEQNPIYWARYADWLFTTPLLLLDLALLVDADQGTILALVGADGIMIGTGLVGALTKVYSYRFVWWAISTAAMLYILYVLFFGFTSKAESMRPEVASTFKVLRNVTVVLWSAYPVVWLIGSEGAGIVPLNIETLLFMVLDVSAKVGFGLILLRSRAIFGEAEAPEPSAGDGAAATS"], PlotLabel -> "window size " <> IntegerString[w], WindowSize -> w], {w, {7, 9, 11}}] // GraphicsColumn](https://www.wolframcloud.com/obj/resourcesystem/images/3a1/3a1da606-52f5-4b25-8b8c-e575fe48c1ca/1c7c8b189a396c2e.png) |
| Out[4]= | 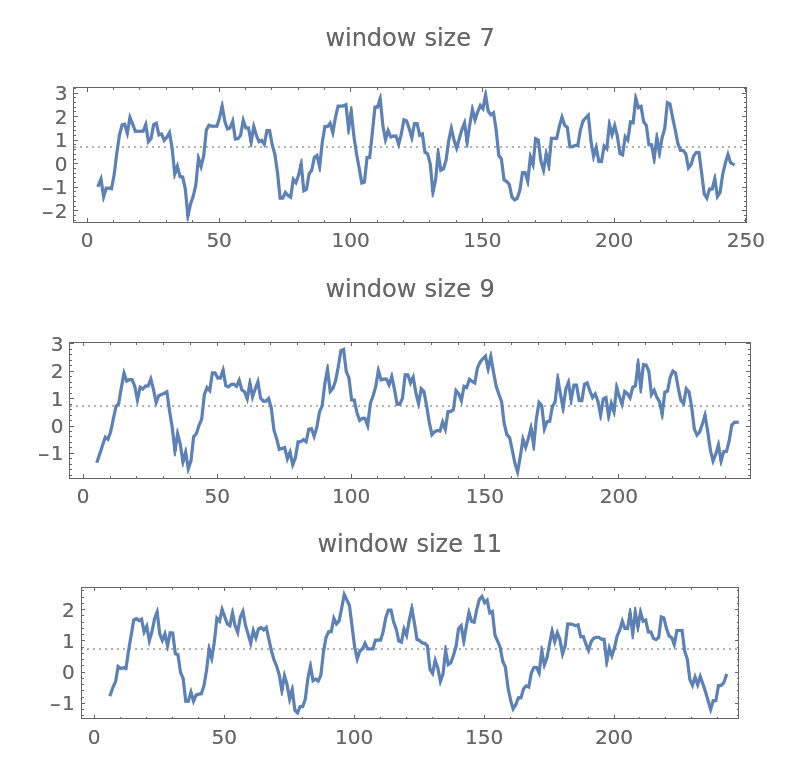 |
Download the amino acid sequence for the protein titin and visualize its hydropathy:
| In[5]:= |
| Out[5]= |  |
This work is licensed under a Creative Commons Attribution 4.0 International License