Details
"Nucleotide" and "Protein" are the supported values for database.
FASTA formatted nucleotide and protein sequences are retrieved from the databases provided by the
NCBI (National Center for Biotechnology Information).
Nucleotide sequences can be queried by their
NCBI Nucleotide Reference Sequence accession number,
GenBank database nucleotide sequence accession number,
RCSB PDB (Research Collaboratory for Structural Bioinformatics Protein Data Bank) accession number.
Protein sequences can be queried by their
NCBI Protein Reference Sequence accession number,
GenBank database protein sequence accession number,
RCSB PDB accession number or
UniProt (Universal Protein Resource) name or accession number.
ResourceFunction["ImportFASTA"][seqref] is equivalent to ResourceFunction["ImportFASTA"][seqref,"Nucleotide"].
The value for format can be any of the following:
| "Data" | "Header" and "Sequence" elements combined in a list |
| "LabeledData" | labeled sequence converted to a rule |
| "FASTA" | FASTA format |
| "BioSequence" | BioSequence format |
ResourceFunction["ImportFASTA"][seqref, database] is equivalent to ResourceFunction["ImportFASTA"][seqref,database,"Data"].





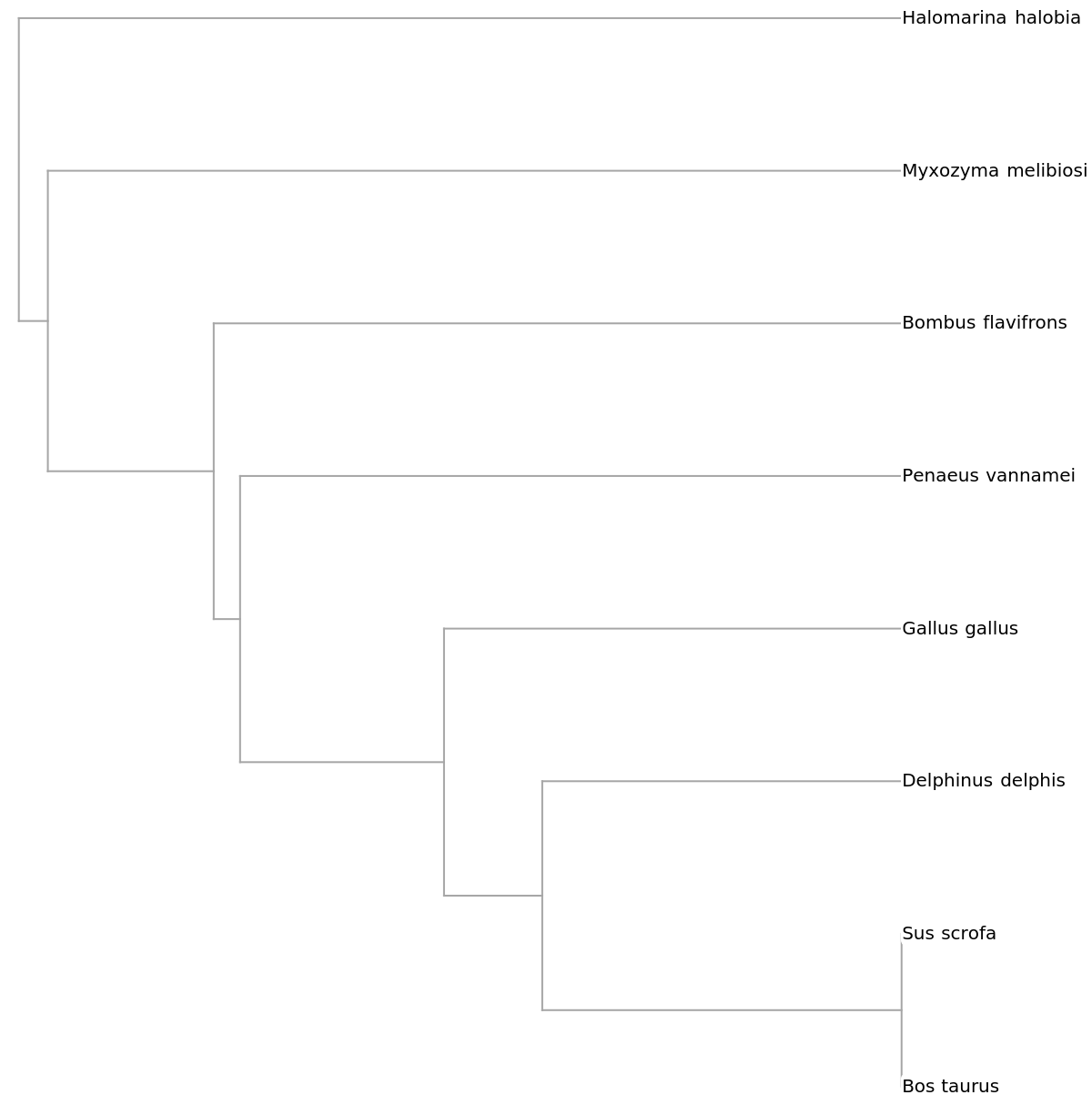
![prot = ResourceFunction["ImportFASTA"][#, "Protein"] & /@ {"NP_001039526.1", "NP_001385227.1", "NP_001123442.1", "XP_069973776.1", "XP_064766840.1", "WP_276305918.1", "XP_059855190.1", "XP_068969445.1"}](https://www.wolframcloud.com/obj/resourcesystem/images/3ca/3ca50e0a-86fb-47da-ba15-8da3cf5eb306/01538c7120af8b49.png)