Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Retrieve virus genome data, including the associated sequence and metadata
ResourceFunction["NCBIVirusGenomeData"][species, "Dataset"] returns the viral sequence dataset for specified species. | |
ResourceFunction["NCBIVirusGenomeData"][species, "Tabular"] returns the viral sequence dataset for specified species in a Tabular format. | |
ResourceFunction["NCBIVirusGenomeData"][species, "Summary"] returns the summary information for the viral dataset of specified species. |
| "APIKey" | None | an API key provided by NCBI; by including a key, up to 10 requests per second are permitted by default |
| "CompleteOnly" | True | limiting to genomes designated as complete, as defined by the submitter. |
| "GeoLocation" | None | limiting to genomes collected from the specified geographic location; entities of the types "Country","GeographicRegion" as well as US states of the type "AdministrativeDivision" are allowed. |
| "Host" | None | limiting to genomes isolated from the specified host species; "TaxonomicSpecies" entity, "NCBITaxonomyID" in an ExternalIdentifier format, or common or scientific name of species is allowed. |
| "IncludeSequence" | None | including specified sequences formatted as BioSequence objects; allowed sequence types include: "Genome", "Protein", "CDS" |
| "PangolinClassification" | None | limiting to SARS-CoV-2 genomes from the specified Pango lineage. |
| "RefSeqOnly" | False | limiting results to RefSeq genomes. |
| "ReleasedSince" | None | limiting to genomes released on or after the specified date. |
| "UpdatedSince" | None | limiting to genomes updated on or after the specified date. |
Retrieve viral genome data for the Zika virus:
| In[1]:= |
| Out[1]= | 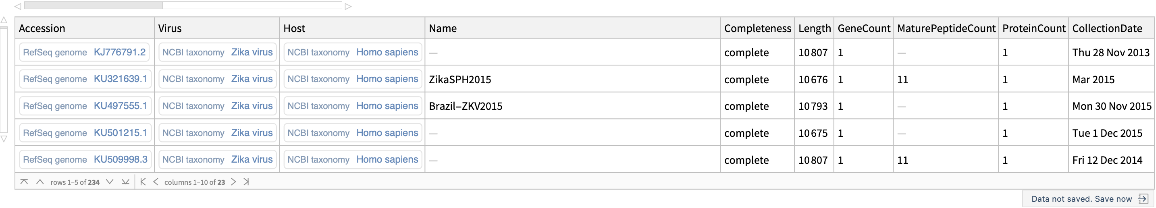 |
Obtain a Tabular form for genome data and protein sequences of the Ebola virus:
| In[2]:= | ![ebola = ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "EbolaVirus::5q9c8"], "Tabular", "IncludeSequence" -> "Protein"]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/740b7ede64c69a5c.png) |
| Out[2]= |  |
Find the size of the dataset for the SARS-CoV-2 genomes:
| In[3]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "SevereAcuteRespiratorySyndromeCoronavirus2::f6fc3"], "Summary", "IncludeSequence" -> "Genome"]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/2c536116c0623e86.png) |
| Out[3]= |  |
Selectively retrieve the SARS-CoV-2 genomes collected in the past two weeks in Connecticut:
| In[4]:= | ![sarscov = ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "SevereAcuteRespiratorySyndromeCoronavirus2::f6fc3"], "ReleasedSince" -> DatePlus[Today, -Quantity[14, "Days"]], "IncludeSequence" -> "Genome", "GeoLocation" -> Entity["AdministrativeDivision", {"Connecticut", "UnitedStates"}]]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/6d52b8851e54069e.png) |
| Out[4]= |  |
Use the "PhylogeneticTreePlot" function to plot a dendrogram for a set of retrieved genome sequences:
| In[5]:= |
| Out[5]= | 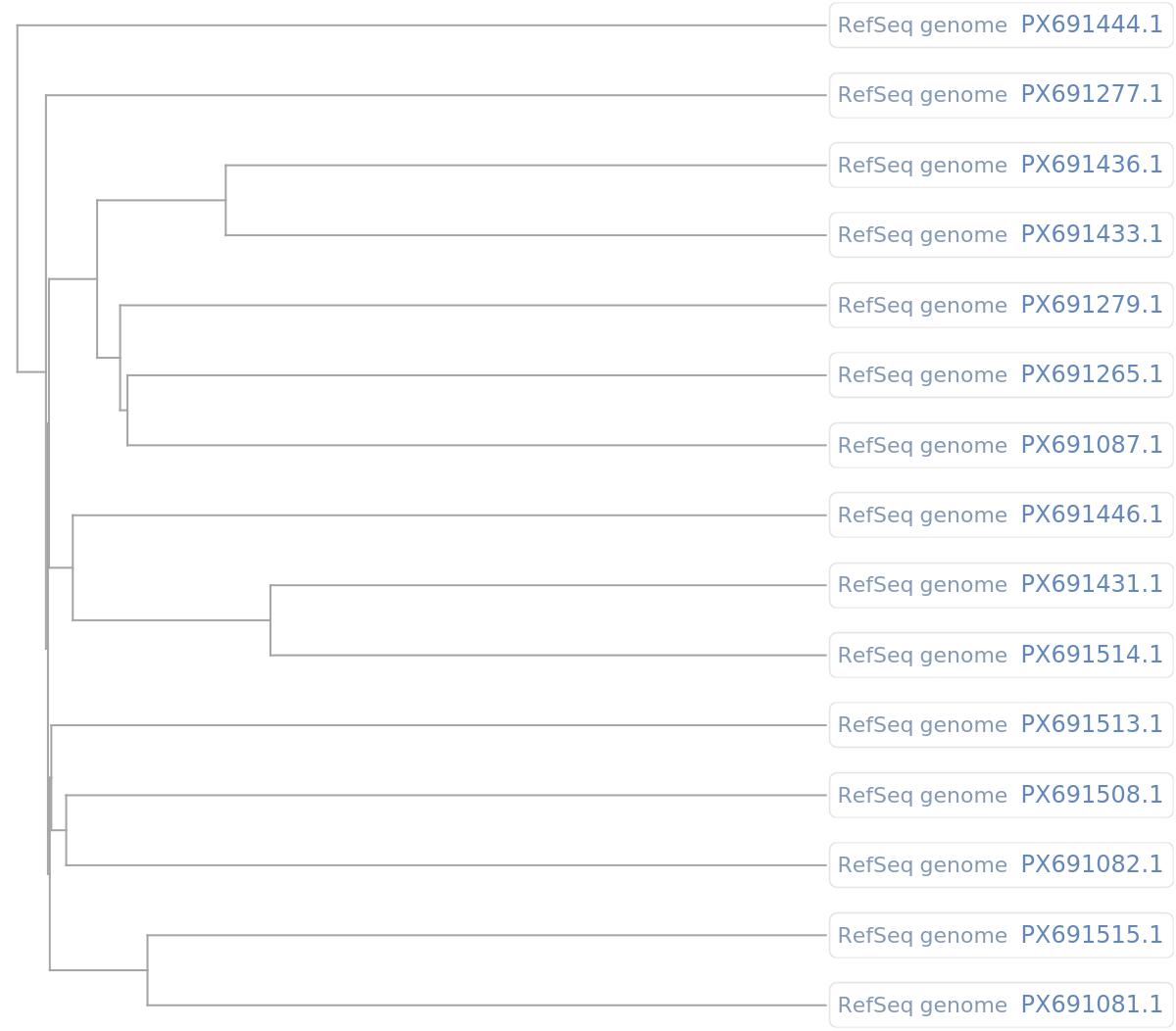 |
Retrieve the Dengue virus genomes:
| In[6]:= |
| Out[6]= |  |
Color countries where genome samples were collected:
| In[7]:= |
| Out[7]= | 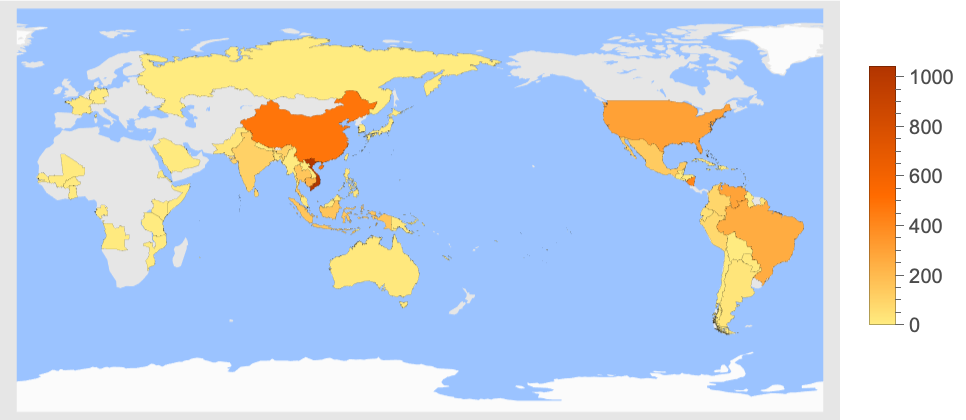 |
Retrieve only the complete genomes:
| In[8]:= |
| Out[8]= |  |
Retrieve the genomes collected in Asia:
| In[9]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "MeaslesMorbillivirus::45qq7"], "GeoLocation" -> Entity["GeographicRegion", "Asia"]]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/0f504dce34c7e297.png) |
| Out[9]= |  |
Retrieve the Tomato yellow leaf curl virus genomes isolated from eggplants:
| In[10]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "TomatoYellowLeafCurlVirus::c36dt"], "Host" -> Entity["TaxonomicSpecies", "SolanumMelongena::9g9tf"]]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/0f09d2ed28d0b3e2.png) |
| Out[10]= | 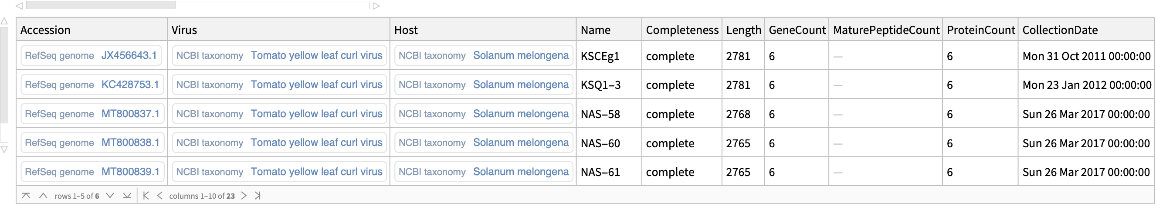 |
Include coding DNA sequences:
| In[11]:= |
| Out[11]= | 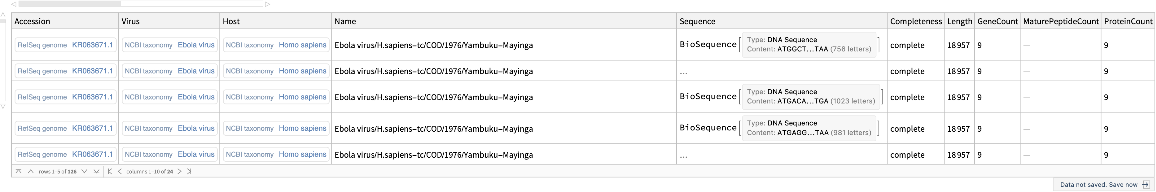 |
Retrieve SARS-CoV-2 genomes from the selected Pango lineage:
| In[12]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "SevereAcuteRespiratorySyndromeCoronavirus2::f6fc3"], "PangolinClassification" -> "LP.8.1"]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/02fed933b319552f.png) |
| Out[12]= |  |
Retrieve the RefSeq genomes:
| In[13]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "HumanAlphaherpesvirus3::6yh3h"], "RefSeqOnly" -> True, "IncludeSequence" -> "Genome"]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/4854779385e99942.png) |
| Out[13]= | 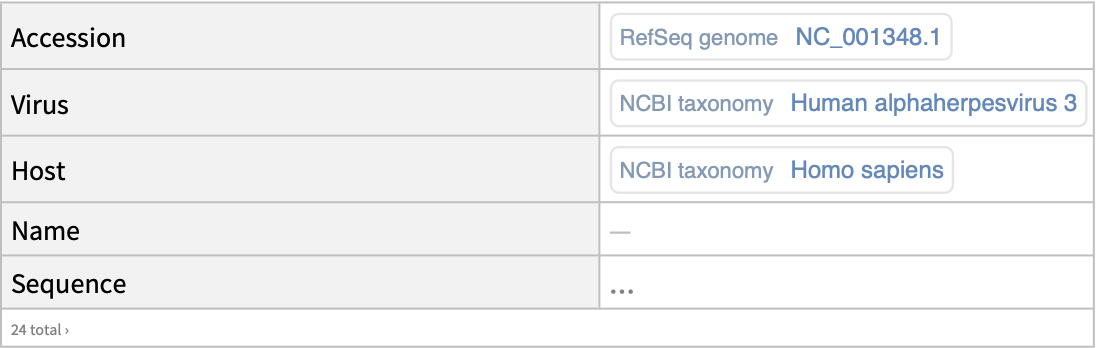 |
Retrieve genomes released in the past year:
| In[14]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "MonkeypoxVirus::9y6ry"], "ReleasedSince" -> DatePlus[Today, -Quantity[1, "Years"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/5788e0c6c67784df.png) |
| Out[14]= | 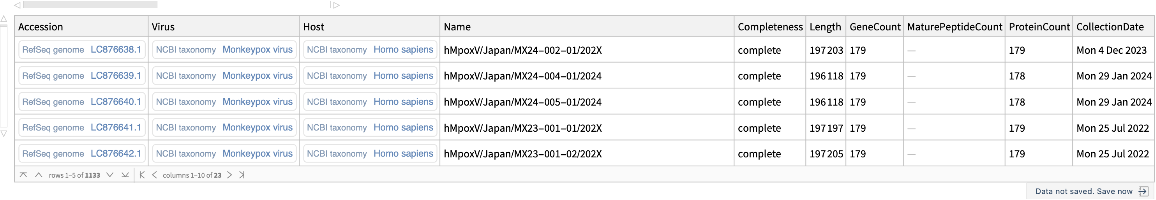 |
Retrieve genomes updated in the past month:
| In[15]:= | ![ResourceFunction["NCBIVirusGenomeData"][
Entity["TaxonomicSpecies", "HepatitisBVirus::rc8p8"], "UpdatedSince" -> DatePlus[Today, -Quantity[1, "Months"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/c8c/c8ce21ad-423a-468d-ab55-db56b96484ab/682289cf28a49515.png) |
| Out[15]= | 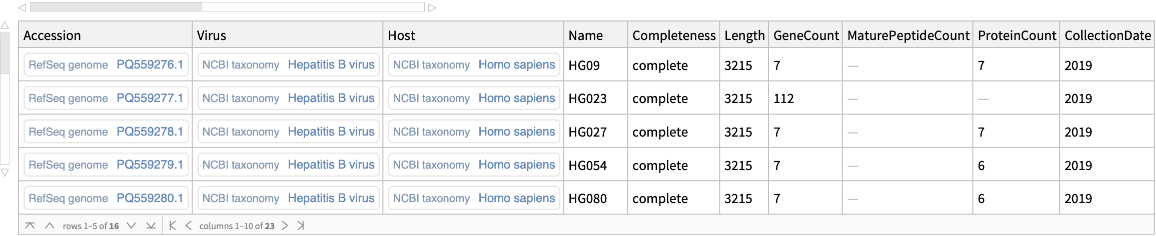 |
Retrieve RefSeq genome data for the Zika virus:
| In[16]:= |
| Out[16]= | 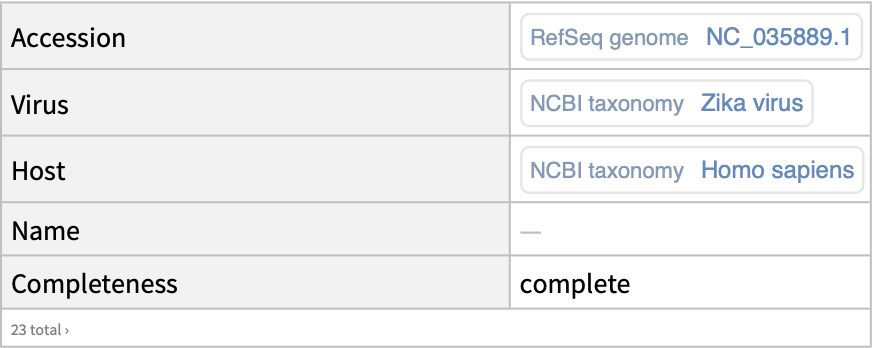 |
Use the "ImportFASTA" function to retrieve the reference sequence:
| In[17]:= |
| Out[17]= |
This work is licensed under a Creative Commons Attribution 4.0 International License