Details and Options
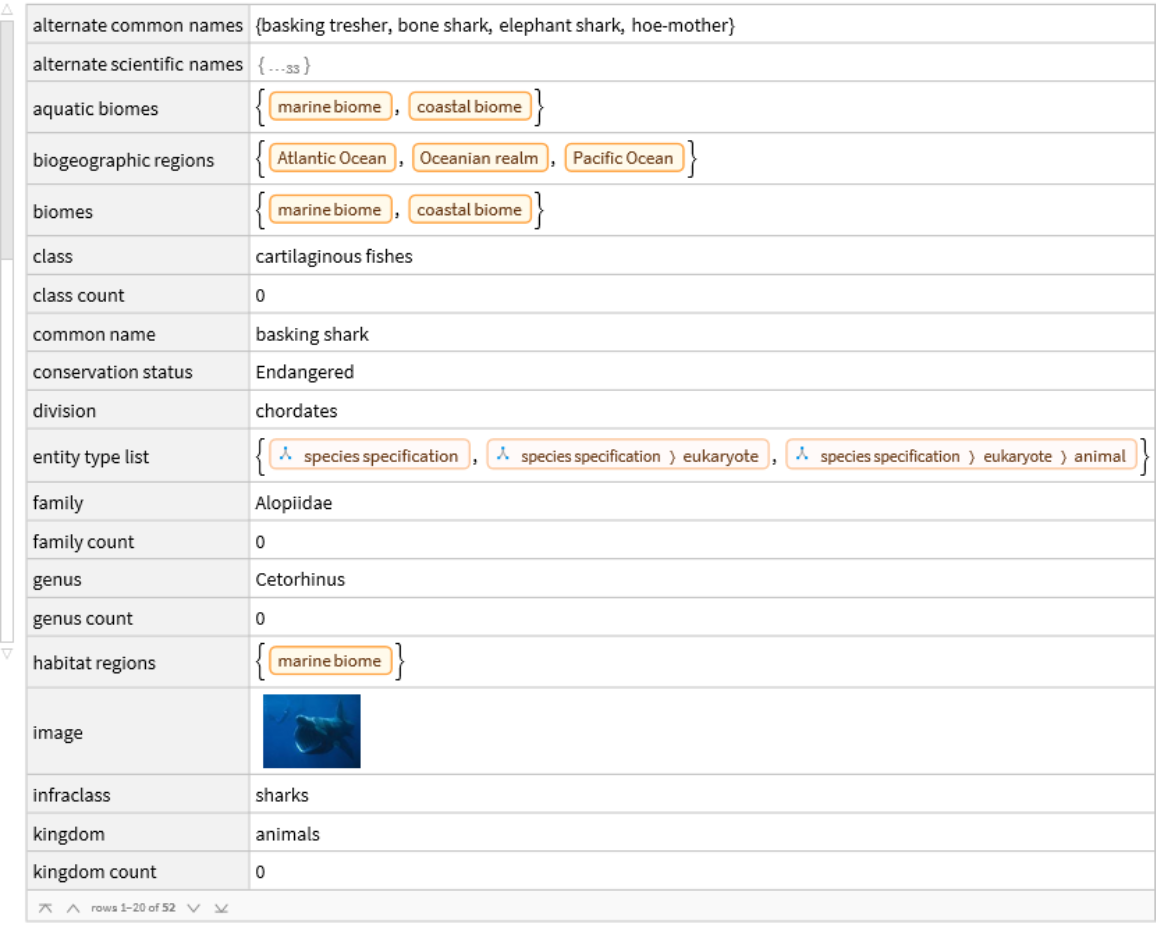
Organisms are classified into a nested hierarchy of taxa, from the broad kingdom and phylum levels down to the specific species level. Each step in the taxonomic sequence represents taxa that belong to the level above and contain subgroups representing the level below.
ResourceFunction["TaxonomyTree"] visualizes the taxonomy of a single or multiple biological organism's entities as a tree.
ResourceFunction["TaxonomyTree"] species arguments are constrained to "TaxonomicSpecies" entities.
Tree element labels are displayed by their
CommonNames. This can be overridden with
Tree Graphics options.
ResourceFunction["TaxonomyTree"] will automatically correct errors in the taxonomic sequence i.e. when multiple edges connect to a single tree element. TaxonomyTree will replace them by one edge, connected to its element's parent taxon.
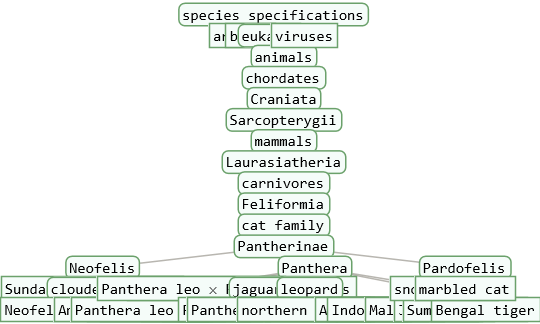
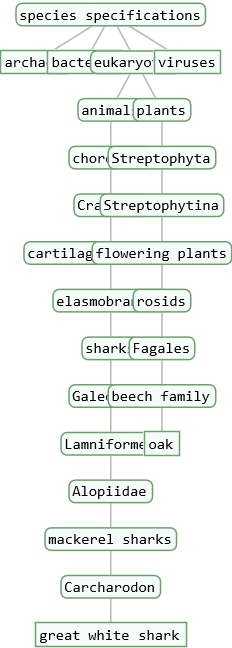
A taxonomy tree of a single entity can be used to study the taxonomy of the species entity in detail. ResourceFunction["TaxonomyTree"] will visualize all nested hierarchies emerging subsequently out of a higher ranking root taxon down to the species and sub-species taxon level.
The number of taxonomic levels between the root- and the species taxon-level can be set with the option "Depth".
The "Depth" option should be a positive integer (default = 3). For example: "Depth" → 4 for the "Species Sapiens" is the Order Primates in the taxonomic sequence: Domain Eukaryotes -> Kingdom Animals -> Phylum Chordates -> Class Mammals -> Order Primates -> Family Hominidae -> Genus Homo -> Species Sapiens.
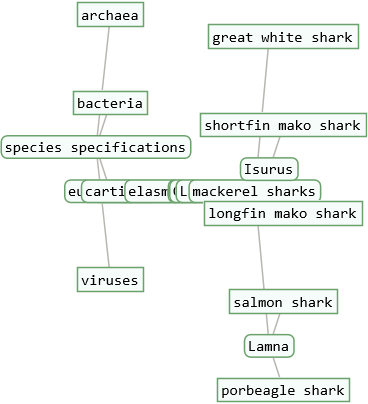
A taxonomy tree of a list of multiple entities can be used to study the taxonomic relationship between entities. ResourceFunction["TaxonomyTree"] will visualize all levels in the taxonomic sequence of each entity in the list, but without their nested hierarchies. Therefore the option "Depth" will be ignored by ResourceFunction["TaxonomyTree"] if a list of entities is used as an argument.

![ResourceFunction["TaxonomyTree"][
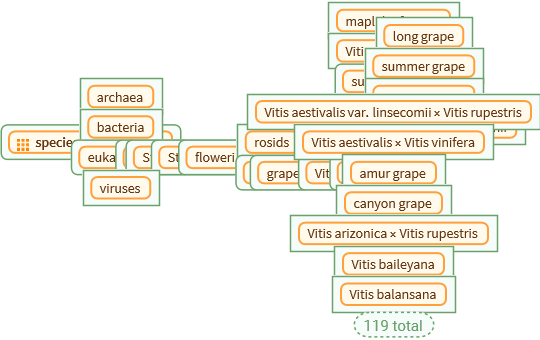
Entity["TaxonomicSpecies", "Vitis::cd68g"], TreeLayout -> Left, "Depth" -> 1, TreeElementLabelFunction -> All -> Automatic]](https://www.wolframcloud.com/obj/resourcesystem/images/45b/45b2ad9d-1e99-4802-abe8-c17809e7d10b/1591e2e5a023e14b.png)

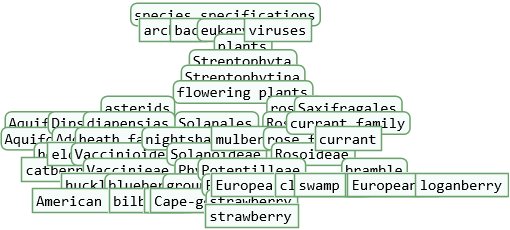
![species = {EntityClass["TaxonomicSpecies", "PlantSpecies"], EntityClass["TaxonomicSpecies", "MammalSpecies"], EntityClass["TaxonomicSpecies", "ArchaeanSpecies"], EntityClass["TaxonomicSpecies", "BacteriumSpecies"], EntityClass["TaxonomicSpecies", "VirusSpecies"]};](https://www.wolframcloud.com/obj/resourcesystem/images/45b/45b2ad9d-1e99-4802-abe8-c17809e7d10b/75f46191f201c993.png)

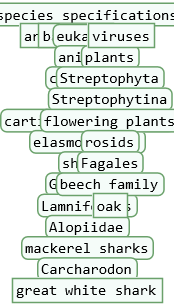
![]() . The tree size at which this download error may occur may vary by species. The error can be prevented by choosing a lower value for the “Depth” option.
. The tree size at which this download error may occur may vary by species. The error can be prevented by choosing a lower value for the “Depth” option.