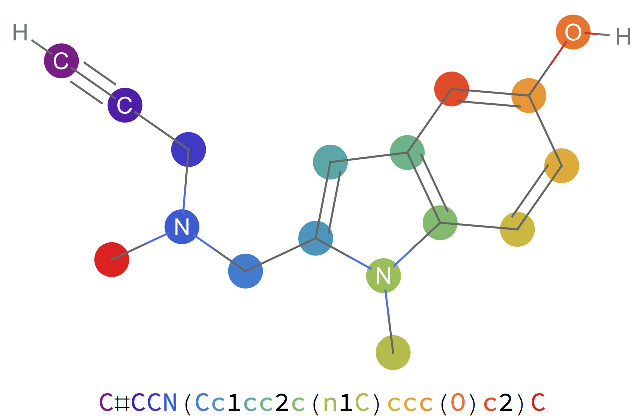
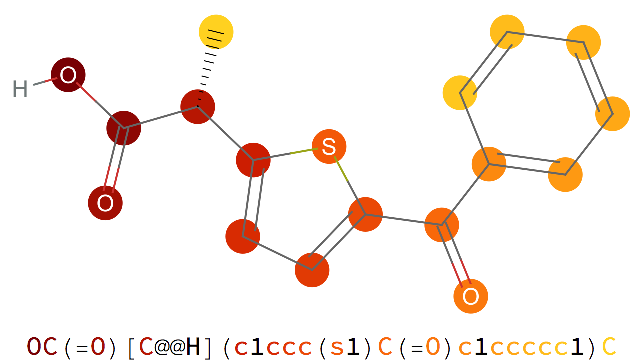
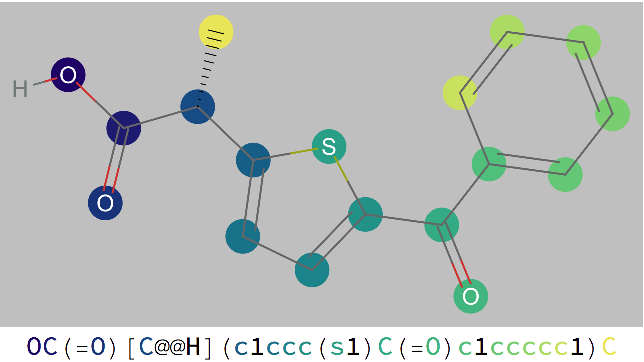
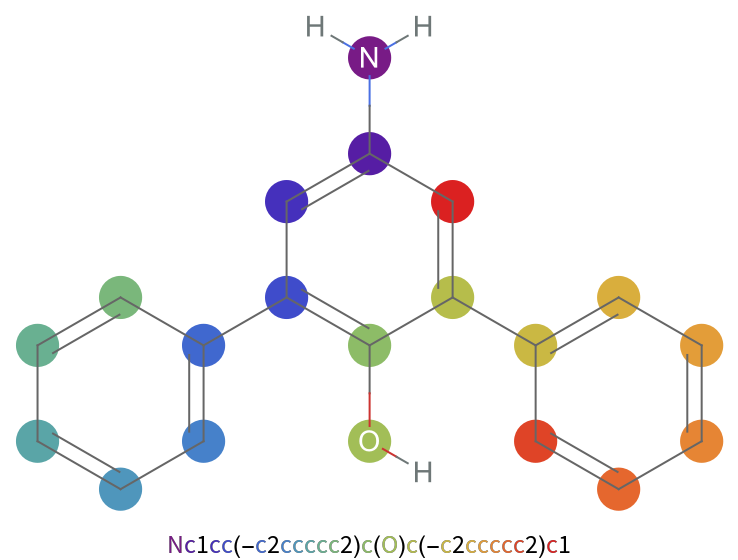
Examples
Basic Examples (2)
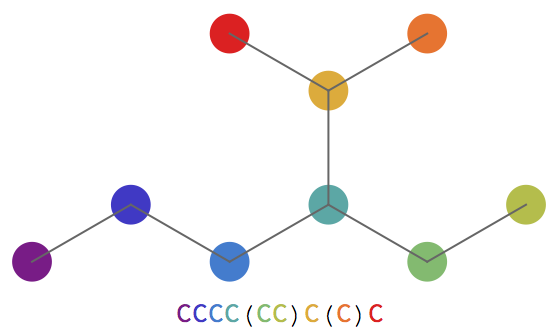
Find the SMILES string for a simple hydrocarbon and visualize it:
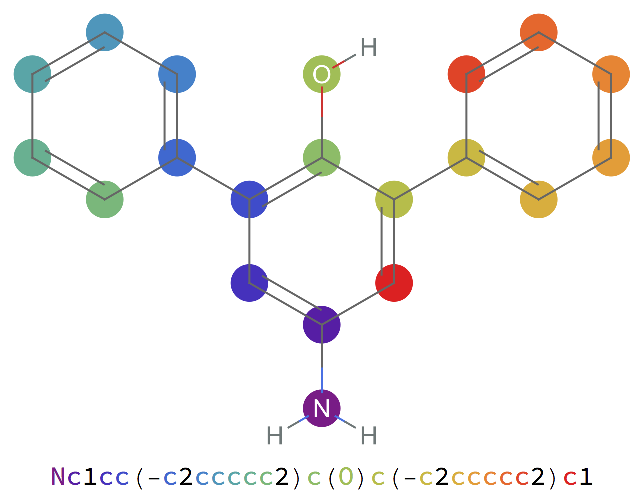
Find the SMILES string for a chemical name and visualize it:
Scope (2)
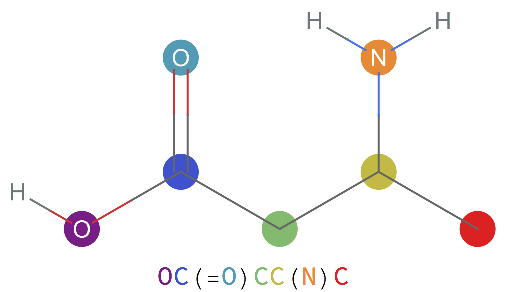
SmilesPlot will work with "Chemical" entities:
SmilesPlot will work with Molecule objects:
Options (2)
Specify a color function for the atoms:
By default, selected atoms are ringed in blue when mousing over the atom code in the SMILES string. Use the option "HighlightColor" to specify an alternate color:
Possible Issues (1)
Some SMILES strings will be canonicalized before plotting:
Publisher
JasonB
Related Links
Version History
-
2.0.0
– 26 April 2023
-
1.0.0
– 05 August 2020
Related Resources
![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/402c90c2-eae8-46bb-bdb3-0ba0874bf47d"]](https://www.wolframcloud.com/obj/resourcesystem/images/8db/8dbdeb82-728f-4ddb-9d51-072e78ae2c49/1-0-0/120318a5957e9f23.png)