Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Use graph algorithms from the Python package NetworkX without any Python programming
ResourceFunction["NetworkXObject"][] returns a configured PythonObject for the Python package NetworkX in a new Python session. | |
ResourceFunction["NetworkXObject"][session] uses the specified running ExternalSessionObject session. | |
ResourceFunction["NetworkXObject"][…,"func"[args,opts]] executes the function func with the specified arguments and options. |
Create a NetworkX object:
| In[1]:= |
| Out[1]= |
Construct an undirected graph from an edge list:
| In[2]:= |
| In[3]:= |
| Out[3]= |
Import the Python object as a Graph:
| In[4]:= |
| Out[4]= | 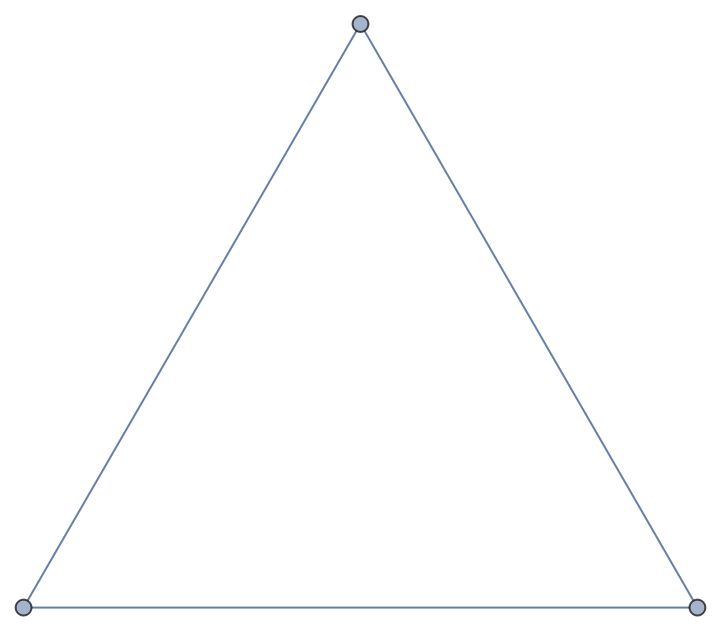 |
The imported graph is a standard Wolfram Language object:
| In[5]:= |
| Out[5]= |
| In[6]:= |
| Out[6]= | 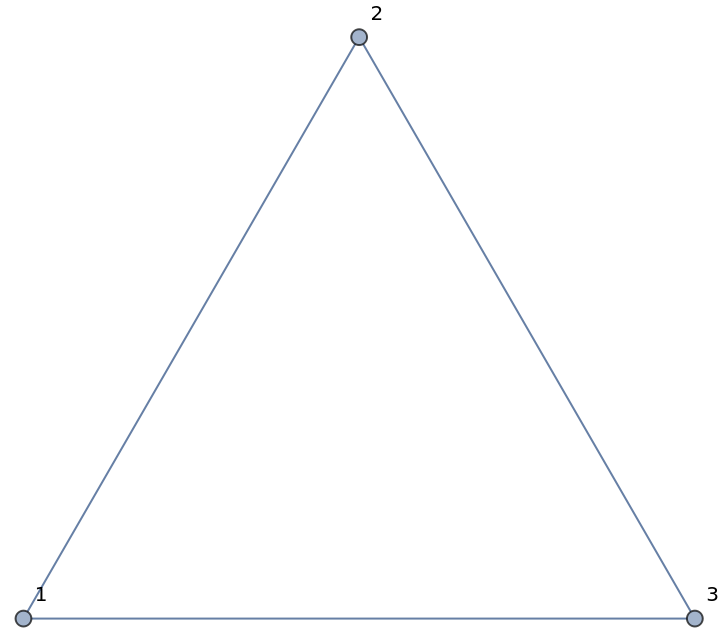 |
Get a directed graph:
| In[7]:= |
| Out[7]= |
| In[8]:= |
| Out[8]= | 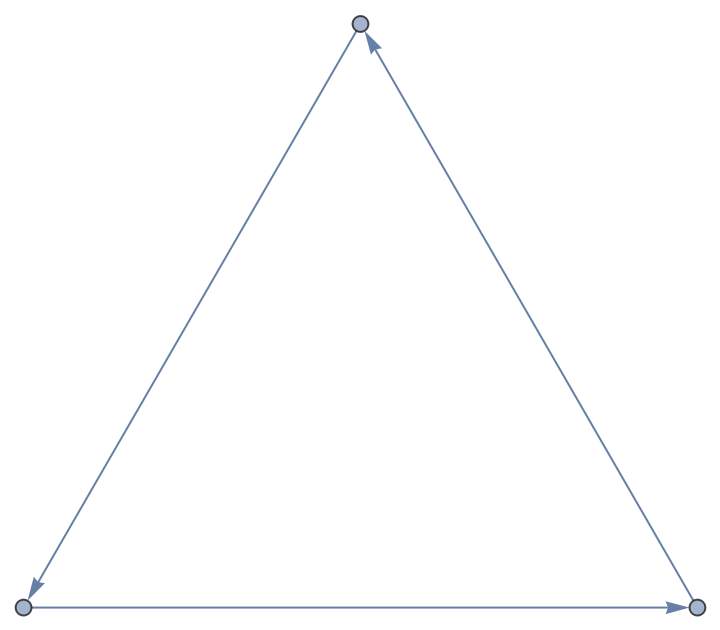 |
| In[9]:= |
| Out[9]= |
Clean up the Python session:
| In[10]:= |
An undirected graph supporting multiple edges between vertices:
| In[11]:= |
| Out[11]= |
| In[12]:= |
| In[13]:= |
| Out[13]= |
| In[14]:= |
| Out[14]= | 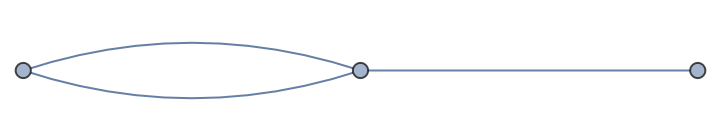 |
| In[15]:= |
| Out[15]= |
A directed multigraph:
| In[16]:= |
| Out[16]= |
| In[17]:= |
| Out[17]= | 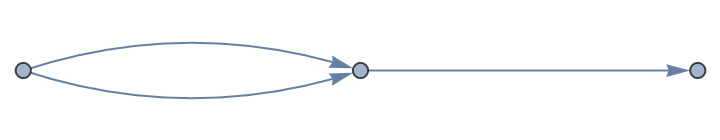 |
| In[18]:= |
| Out[18]= |
| In[19]:= |
Create an empty graph:
| In[20]:= |
| Out[20]= |
| In[21]:= |
| Out[21]= |
Nodes and edges of the graph are given as continuously updated NodeView and EdgeView objects:
| In[22]:= |
| Out[22]= | 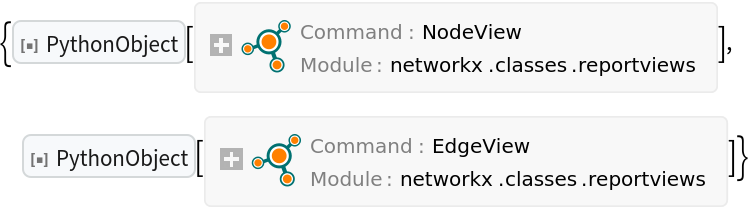 |
| In[23]:= |
| Out[23]= |
Add one node at a time:
| In[24]:= |
Examine nodes:
| In[25]:= |
| Out[25]= |
| In[26]:= |
| Out[26]= |
Remove a node:
| In[27]:= |
| In[28]:= |
| Out[28]= |
Add nodes from a list:
| In[29]:= |
| In[30]:= |
| Out[30]= |
Remove several nodes:
| In[31]:= |
| In[32]:= |
| Out[32]= |
| In[33]:= |
| In[34]:= |
| Out[34]= |
| In[35]:= |
| Out[35]= |
Add nodes along with node attributes specified as an Association:
| In[36]:= |
| In[37]:= |
| Out[37]= |
Retrieve the specified node attribute:
| In[38]:= |
| Out[38]= |
Check the attributes of the imported graph:
| In[39]:= |
| Out[39]= |  |
| In[40]:= |
| Out[40]= |
| In[41]:= |
| Out[41]= |
| In[42]:= |
Create a new graph:
| In[43]:= |
| Out[43]= |
| In[44]:= |
| Out[44]= |
To add a node attribute after a node is created, get a reference to the Python dictionary of node attributes:
| In[45]:= |
| In[46]:= |
| Out[46]= |  |
Adding a key→value pair to the dictionary adds an attribute to the graph:
| In[47]:= |
| Out[47]= |
| In[48]:= |
| Out[48]= |
| In[49]:= |
Create a new graph:
| In[50]:= |
| Out[50]= |
| In[51]:= |
| Out[51]= |
Grow the graph by adding one edge at a time:
| In[52]:= |
| In[53]:= |
| Out[53]= |
Add a list of edges:
| In[54]:= |
Add a list of edges with attributes:
| In[55]:= |
| In[56]:= |
| Out[56]= |
Check the attributes in the imported graph:
| In[57]:= |
| Out[57]= | 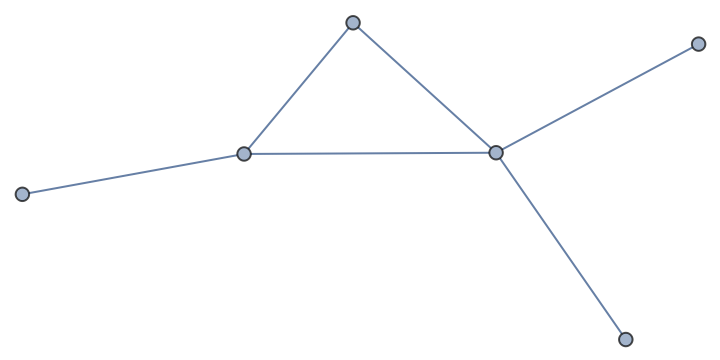 |
| In[58]:= |
| Out[58]= |
| In[59]:= |
| Out[59]= |
To add an edge attribute after an edge is created, assign a key→value pair to the dictionary of edge attributes:
| In[60]:= |
| Out[60]= |  |
| In[61]:= |
| Out[61]= |
Confirm the assignment for the edge 12:
| In[62]:= |
| Out[62]= |
Remove a single edge:
| In[63]:= |
| In[64]:= |
| Out[64]= |
Remove several edges:
| In[65]:= |
| In[66]:= |
| Out[66]= |
| In[67]:= |
Create a new empty graph:
| In[68]:= |
| Out[68]= |
Incorporate nodes from one graph into another:
| In[69]:= |
| Out[69]= | 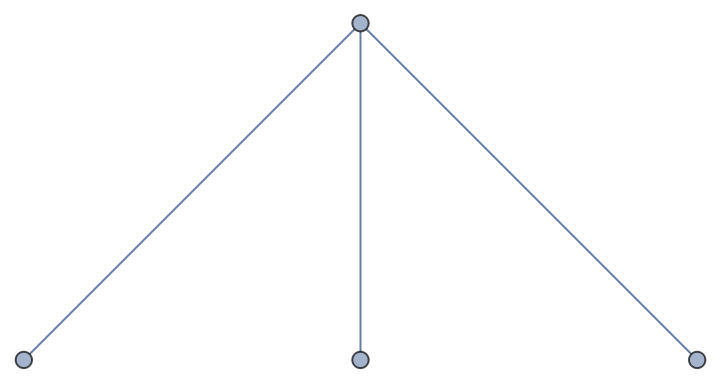 |
| In[70]:= |
| Out[70]= |
| In[71]:= |
| In[72]:= |
| Out[72]= |
| In[73]:= |
| Out[73]= | 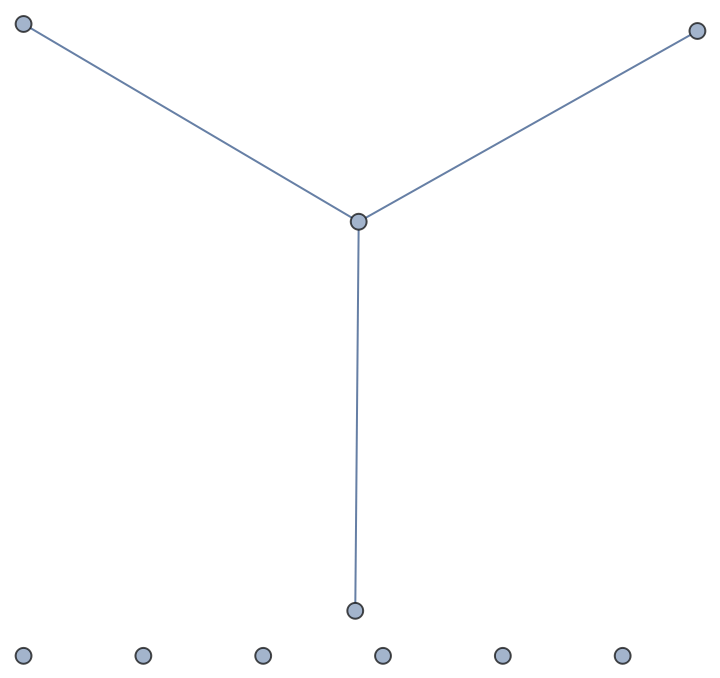 |
| In[74]:= |
Create a new empty graph:
| In[75]:= |
| Out[75]= |
Use one graph as a node in another:
| In[76]:= |
| Out[76]= |
| In[77]:= |
| Out[77]= |
| In[78]:= |
| In[79]:= |
| Out[79]= |
| In[80]:= |
| Out[80]= | 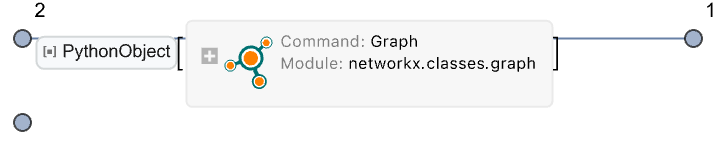 |
| In[81]:= |
Create a new empty graph:
| In[82]:= |
| Out[82]= |
| In[83]:= |
| Out[83]= |
Add a node "spam":
| In[84]:= |
Add four nodes "s", "p", "a", "m":
| In[85]:= |
| In[86]:= |
| Out[86]= |
| In[87]:= |
Create a new empty graph:
| In[88]:= |
| Out[88]= |
| In[89]:= |
| Out[89]= |
Incorporate edges from one graph into another:
| In[90]:= |
| Out[90]= | 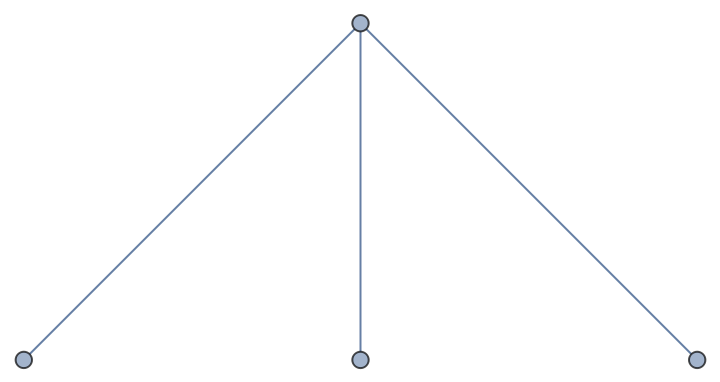 |
| In[91]:= |
| Out[91]= |
| In[92]:= |
| In[93]:= |
| Out[93]= |
| In[94]:= |
| Out[94]= | 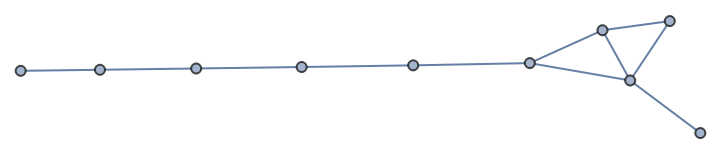 |
| In[95]:= |
Create a new multi-digraph graph:
| In[96]:= |
| Out[96]= |
| In[97]:= |
| Out[97]= |
Add edges with weights:
| In[98]:= |
| In[99]:= |
| Out[99]= |
| In[100]:= |
| Out[100]= |
| In[101]:= |
Create a new multi-digraph:
| In[102]:= |
| Out[102]= |
Count nodes and edges in a graph:
| In[103]:= |
| Out[103]= |
| In[104]:= |
| Out[104]= | 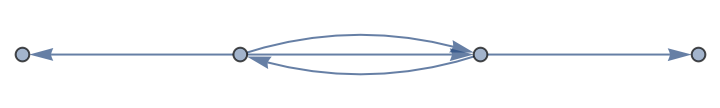 |
| In[105]:= |
| Out[105]= |
| In[106]:= |
| Out[106]= |
| In[107]:= |
Create a new multi-digraph:
| In[108]:= |
| Out[108]= |
| In[109]:= |
| Out[109]= |
| In[110]:= |
| Out[110]= | 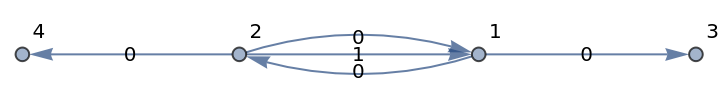 |
Test whether a node is in the graph:
| In[111]:= |
| Out[111]= |  |
| In[112]:= |
| Out[112]= |
Test if an edge is in the graph:
| In[113]:= |
| Out[113]= |
Test if an edge with a specific tag is in the graph:
| In[114]:= |
| Out[114]= |
Use the DirectedEdge wrapper in the edge specification:
| In[115]:= |
| Out[115]= |
| In[116]:= |
Create a new multi-digraph:
| In[117]:= |
| Out[117]= |
| In[118]:= |
| Out[118]= |
Get an Association of neighbors (adjacencies) as a property of the graph:
| In[119]:= |
| Out[119]= |
| In[120]:= |
| Out[120]= |
Alternatively, use the "Adjacency" method to get a list of tuples rather than an Association:
| In[121]:= |
| Out[121]= |  |
| In[122]:= |
| Out[122]= |
| In[123]:= |
Create a new multi-digraph:
| In[124]:= |
| Out[124]= |
| In[125]:= |
| Out[125]= |
Get an object containing vertex degrees for all vertices in the graph:
| In[126]:= |
| Out[126]= |
| In[127]:= |
| Out[127]= |
The degree of a specific vertex:
| In[128]:= |
| Out[128]= |
| In[129]:= |
Create a new multi-digraph:
| In[130]:= |
| Out[130]= |
| In[131]:= |
| Out[131]= |
Get a list of neighbors for a node:
| In[132]:= |
| Out[132]= |  |
| In[133]:= |
| Out[133]= |
Or equivalently, for a directed graph:
| In[134]:= |
| Out[134]= |
Predecessors of a node of a directed graph:
| In[135]:= |
| Out[135]= |
| In[136]:= |
Create a new graph:
| In[137]:= |
| Out[137]= |
| In[138]:= |
| Out[138]= |
Use the special attribute "weight" to create a waited graph:
| In[139]:= |
Alternatively, use the AddWeightedEdgesFrom method:
| In[140]:= |
Compute the weighted adjacency matrix on the Python side:
| In[141]:= |
| Out[141]= |  |
Import the matrix as SparseArray:
| In[142]:= |
| Out[142]= |
| In[143]:= |
| Out[143]= |
The imported graph is weighted too:
| In[144]:= |
| Out[144]= | 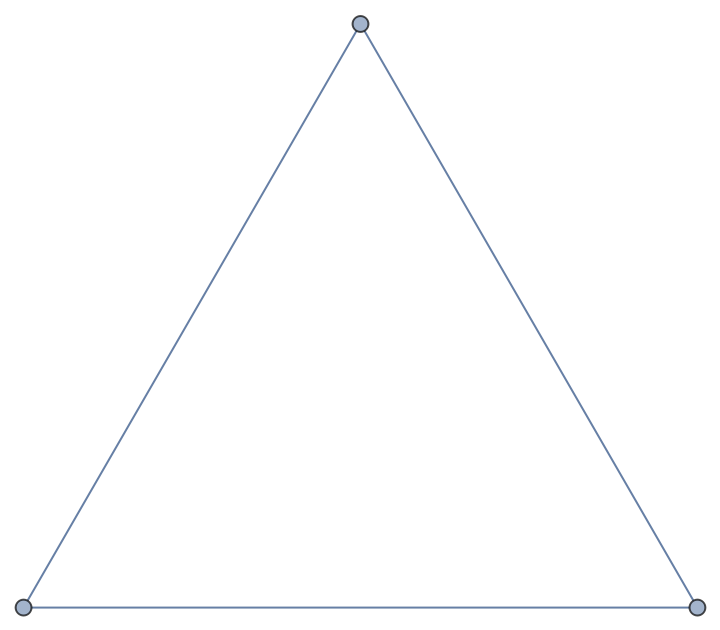 |
| In[145]:= |
| Out[145]= |
| In[146]:= |
| Out[146]= |
| In[147]:= |
| Out[147]= |
Compare with the adjacency matrix computed in Python:
| In[148]:= |
| Out[148]= |
| In[149]:= |
Create a new NetworkX object:
| In[150]:= |
| Out[150]= |
| In[151]:= |
Undirected and directed graphs with self-loops and a single edge between nodes:
| In[152]:= |
| Out[152]= |  |
| In[153]:= |
| Out[153]= | 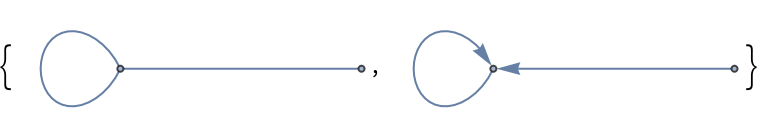 |
| In[154]:= |
| Out[154]= |
Undirected and directed graphs possibly with parallel edges between nodes:
| In[155]:= |
| Out[155]= |  |
| In[156]:= |
| Out[156]= | 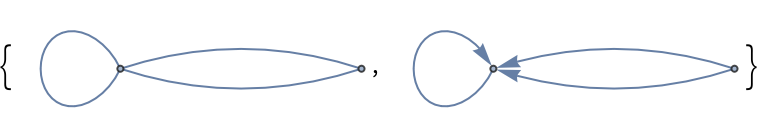 |
| In[157]:= |
| Out[157]= |
Edges in imported multigraphs are tagged:
| In[158]:= |
| Out[158]= |
| In[159]:= |
Build a graph incrementally:
| In[160]:= |
| Out[160]= |
| In[161]:= |
| In[162]:= |
| In[163]:= |
| Out[163]= |  |
| In[164]:= |
Create from an edge list:
| In[165]:= |
| Out[165]= |
Alternatively, use the edge list in the form returned by EdgeList:
| In[166]:= |
| Out[166]= |
| In[167]:= |
| Out[167]= |
| In[168]:= |
Form an Association mapping nodes to neighbors:
| In[169]:= |
| Out[169]= |
| In[170]:= |
| Out[170]= |  |
| In[171]:= |
Form a general Wolfram Language Graph:
| In[172]:= |
| Out[172]= | 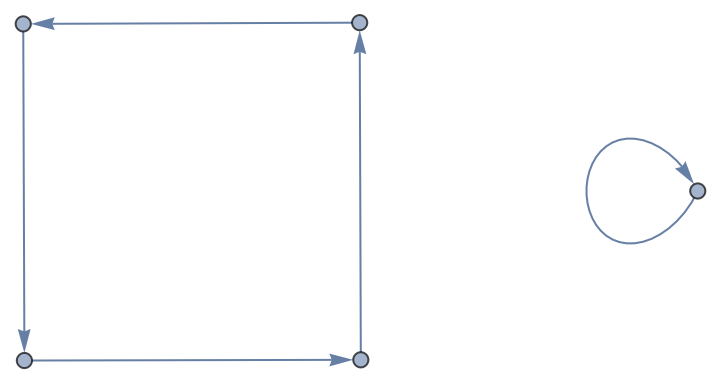 |
| In[173]:= |
| Out[173]= | 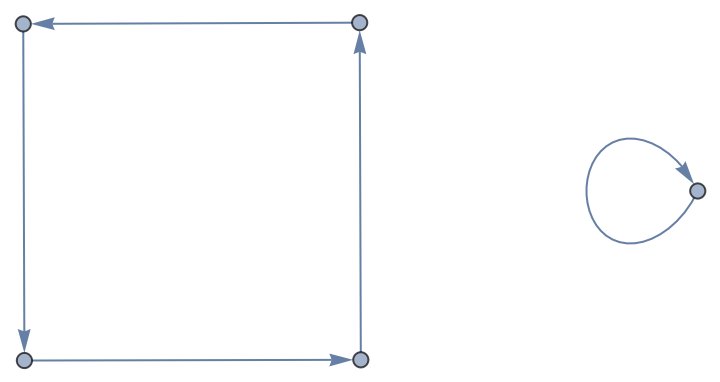 |
From a sparse matrix graph:
| In[174]:= |
| Out[174]= |
| In[175]:= |
| Out[175]= | 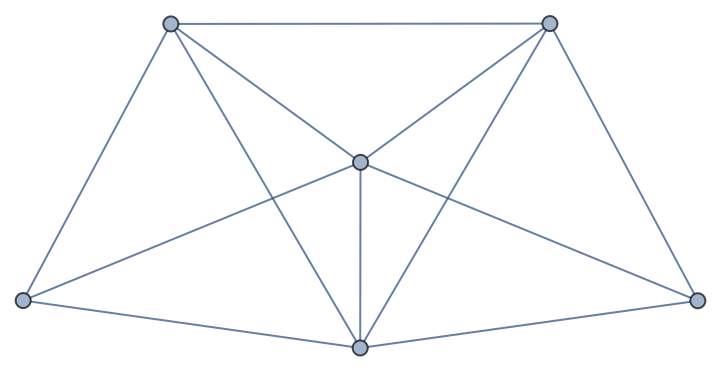 |
| In[176]:= |
| Out[176]= | 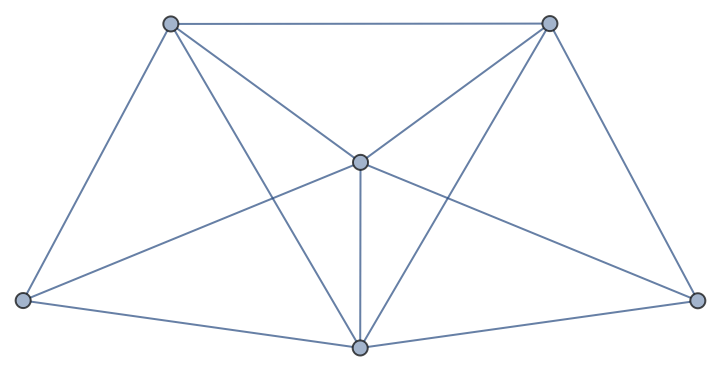 |
| In[177]:= |
From a weighted Graph object:
| In[178]:= |
| In[179]:= |
| Out[179]= | 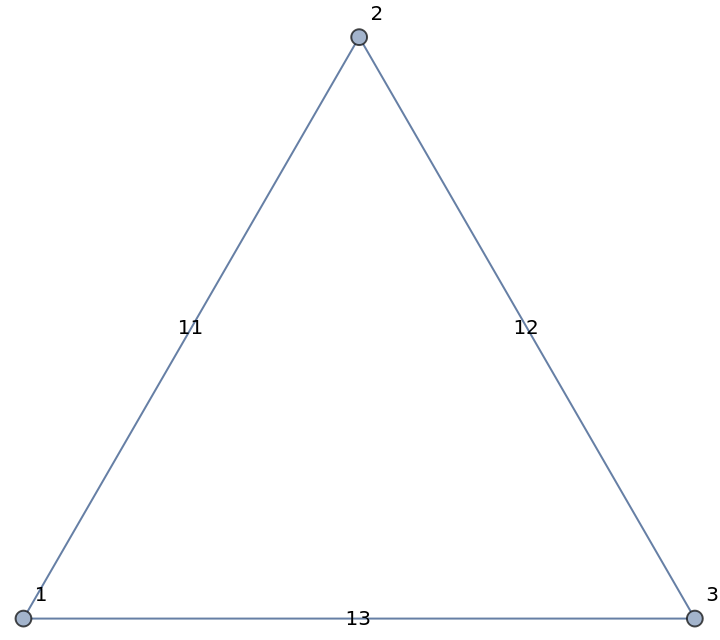 |
| In[180]:= |
| Out[180]= |
| In[181]:= |
| Out[181]= | 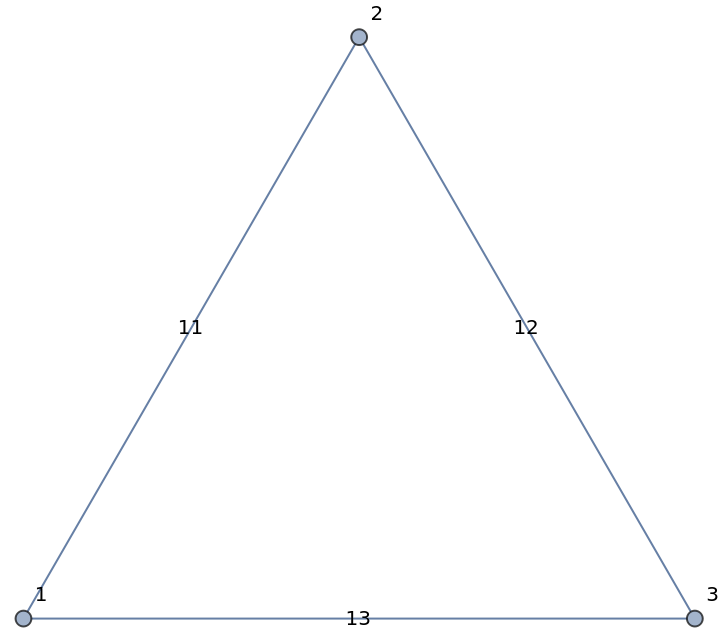 |
| In[182]:= |
From an edge-capacity and vertex-capacity Graph object:
| In[183]:= |
| Out[183]= | 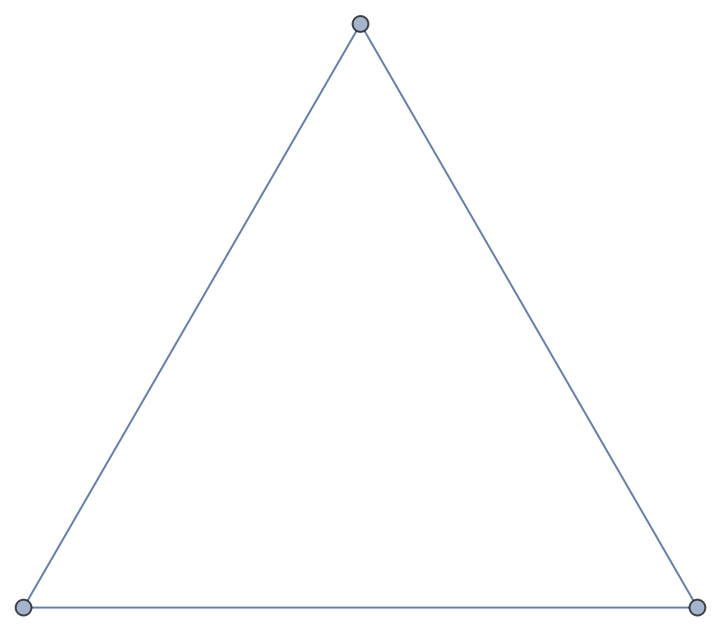 |
Send the graph through NetworkX:
| In[184]:= |
| Out[184]= |
| In[185]:= |
| Out[185]= | 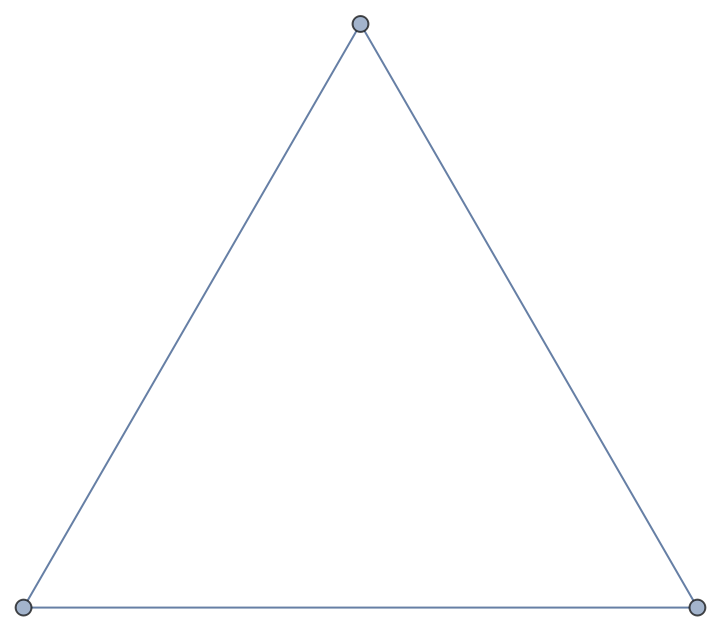 |
The capacity is preserved as an annotation:
| In[186]:= |
| Out[186]= |
| In[187]:= |
| Out[187]= |
| In[188]:= |
From a multigraph:
| In[189]:= |
| In[190]:= |
| Out[190]= |
| In[191]:= |
| Out[191]= |  |
| In[192]:= |
From another NetworkX graph:
| In[193]:= |
| Out[193]= | 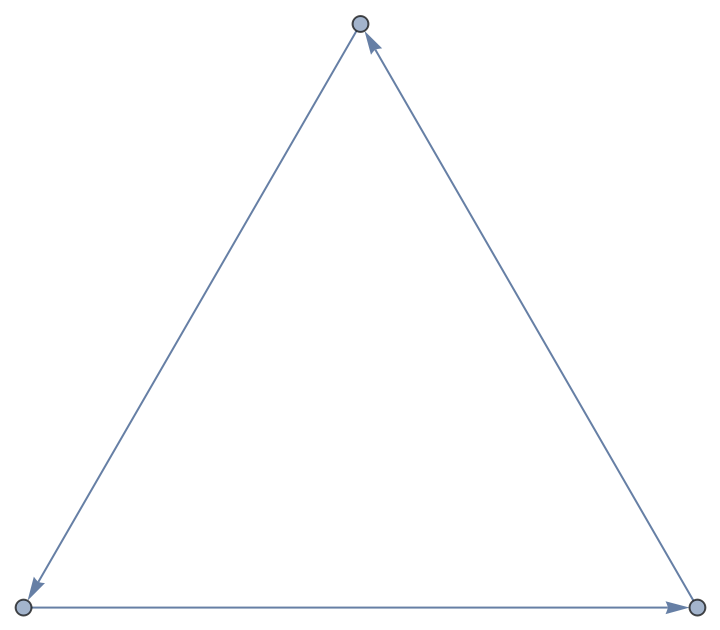 |
| In[194]:= |
| Out[194]= | 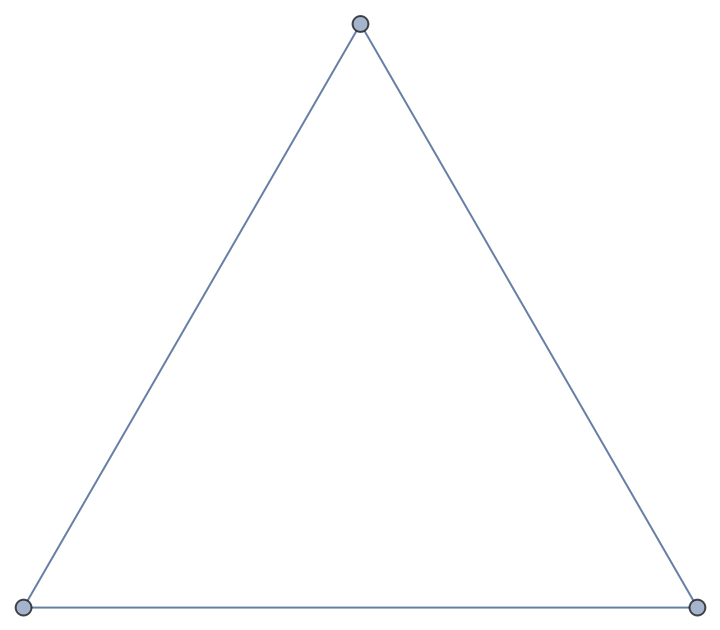 |
| In[195]:= |
Form a 2D NumPy array:
| In[196]:= |
| Out[196]= |
Create a graph and import it:
| In[197]:= | ![nx = ResourceFunction["NetworkXObject"][];
np = ResourceFunction["PythonObject"][
nx["Session"], <|"Command" -> "``", "TemplateArguments" -> {arr}|>]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/5f9bc78b305638ac.png) |
| Out[197]= |  |
| In[198]:= |
| Out[198]= | 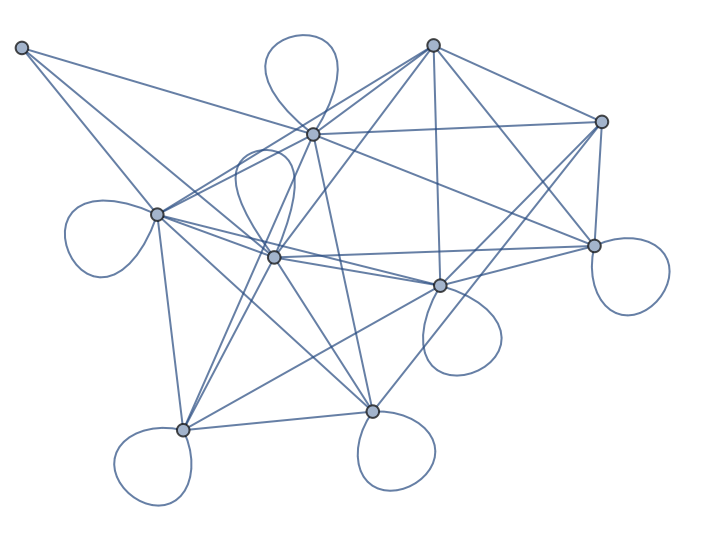 |
The same NumPy array can be supplied implicitly, as long as the array is in the form of NumericArray:
| In[199]:= |
| Out[199]= | 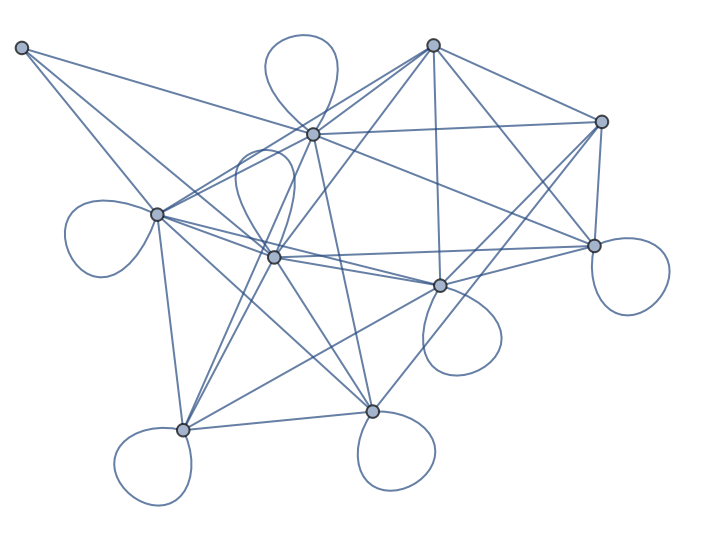 |
| In[200]:= |
Create a graph from a SciPy sparse matrix:
| In[201]:= |
| Out[201]= |  |
| In[202]:= |
| Out[202]= |
| In[203]:= |
| Out[203]= |  |
| In[204]:= |
| Out[204]= | 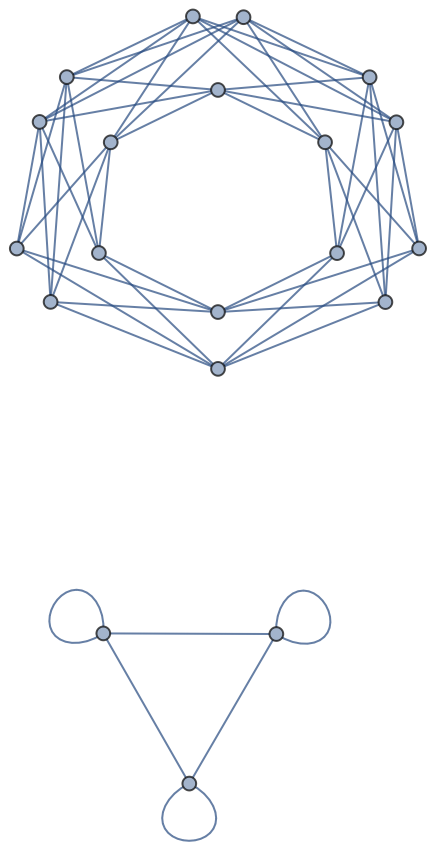 |
Or from a SciPy sparse array:
| In[205]:= |
| Out[205]= |  |
| In[206]:= |
| Out[206]= | 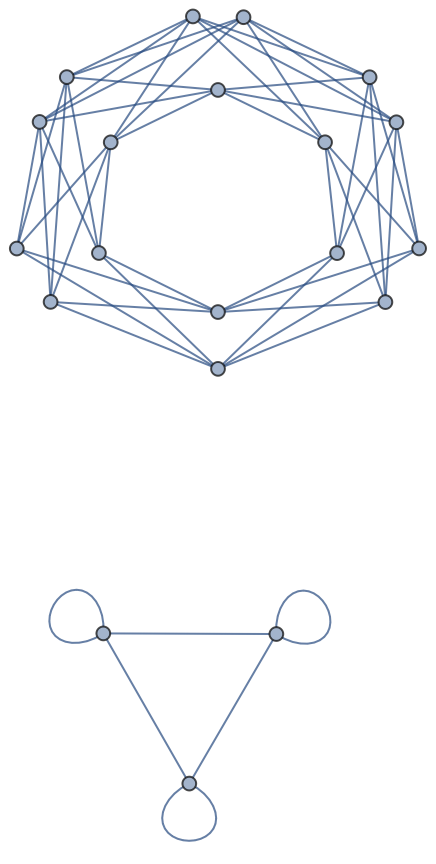 |
| In[207]:= |
Create a graph directly from a SparseArray object:
| In[208]:= | ![s = SparseArray[{Band[{2, 1}] -> 1, Band[{3, 1}] -> 1}, {10, 10}];
nx = ResourceFunction["NetworkXObject"][];
Normal[nx["Graph"[s]]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/61199b7dac2171f0.png) |
| Out[208]= |  |
| In[209]:= |
Create a data frame from a pandas object:
| In[210]:= | ![nx = ResourceFunction["NetworkXObject"][];
pd = ResourceFunction["PandasObject"][nx["Session"]];
df = pd["DataFrame"[{{1, 1}, {2, 1}}]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/317ba91b0988dd6f.png) |
| Out[208]= |
Create a graph:
| In[211]:= |
| Out[211]= |
Or use the FromPandasAdjacency function:
| In[212]:= |
| Out[212]= |
| In[213]:= |
| Out[213]= | 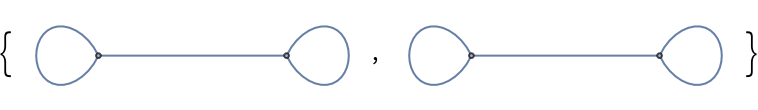 |
| In[214]:= |
Create a data frame representing a pandas edge list:
| In[215]:= | ![nx = ResourceFunction["NetworkXObject"][];
pd = ResourceFunction["PandasObject"][nx["Session"]];
df = pd["DataFrame"[<|"source" -> {0, 1, 2, 0}, "target" -> {2, 2, 3, 2}, "my_edge_key" -> {"A", "B", "C", "D"}, "weight" -> {3, 4, 5, 6}, "color" -> {"red", "blue", "blue", "blue"}|>]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/7b98d5976bb9af36.png) |
| Out[208]= |
Convert to a graph:
| In[216]:= | ![g = nx["FromPandasEdgeList"[
df,
"EdgeKey" -> "my_edge_key",
"EdgeAttr" -> {"weight", "color"},
"CreateUsing" -> nx["MultiGraph"[]]
]
]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/080cd958aa1201e9.png) |
| Out[216]= |
See the Graph:
| In[217]:= |
| Out[217]= |  |
See the edge list and weights:
| In[218]:= |
| Out[218]= | 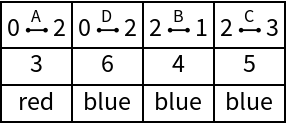 |
Compare with a dataset:
| In[219]:= |
| Out[219]= | 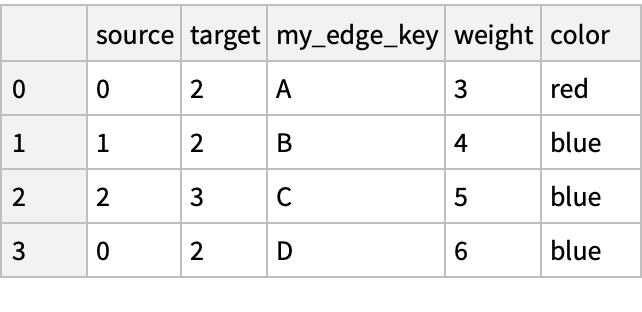 |
| In[220]:= |
Explore the variety of ways to generate graphs:
| In[221]:= |
| In[222]:= |
| Out[222]= | 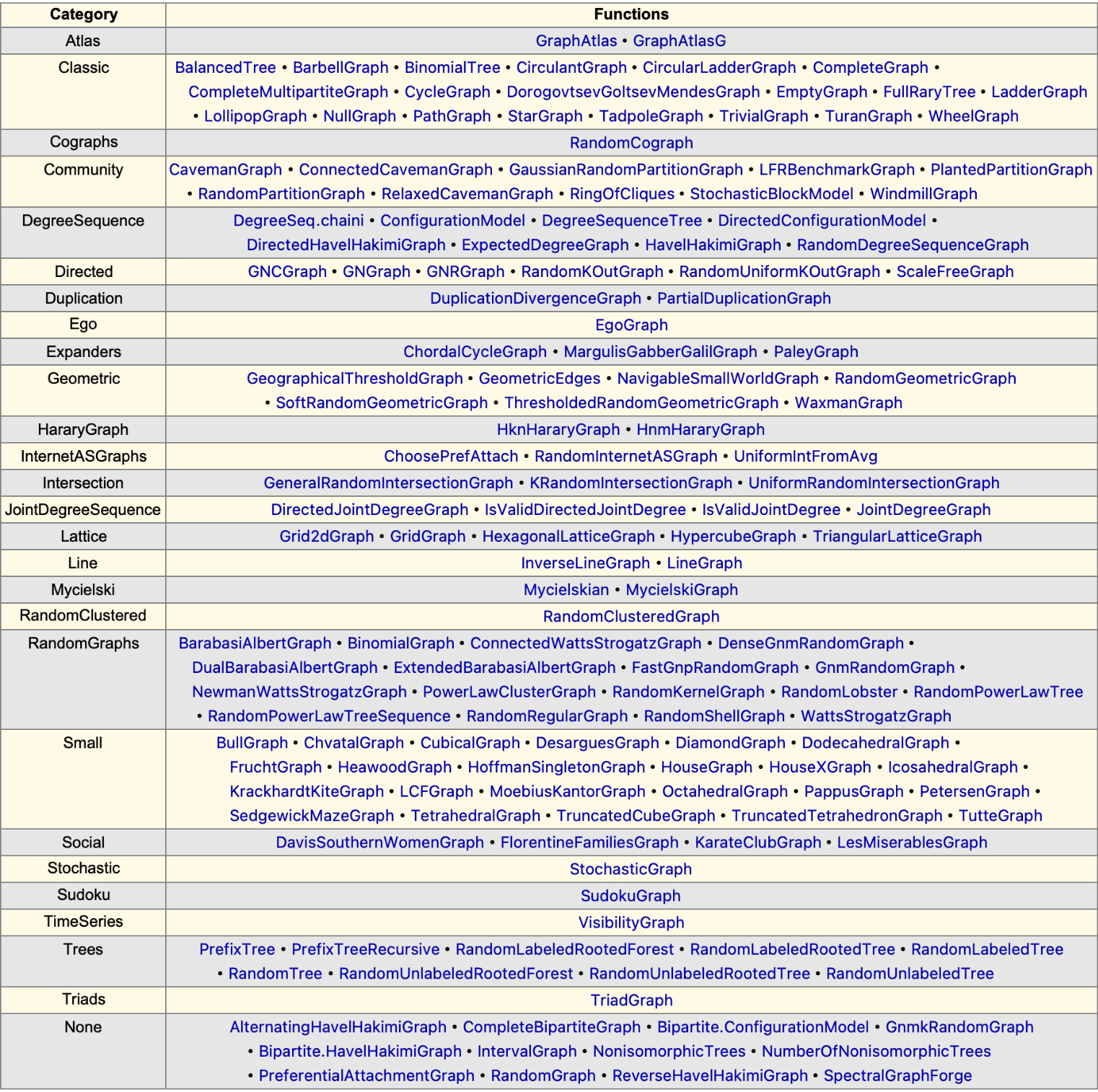 |
| In[223]:= |
Create the first few scale-free pseudo-fractal graphs using the Dorogovtsev-Goltsev-Mendes algorithm:
| In[224]:= |
| Out[224]= | 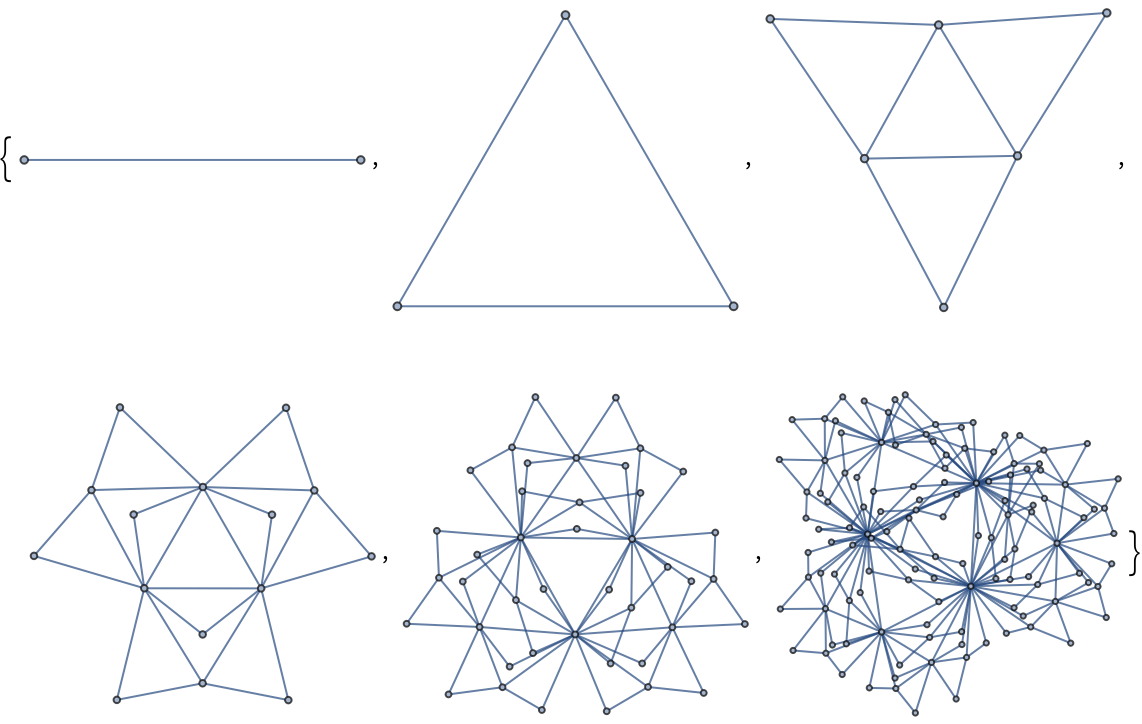 |
| In[225]:= |
A few Newman–Watts–Strogatz small-world graphs:
| In[226]:= |
| Out[226]= | 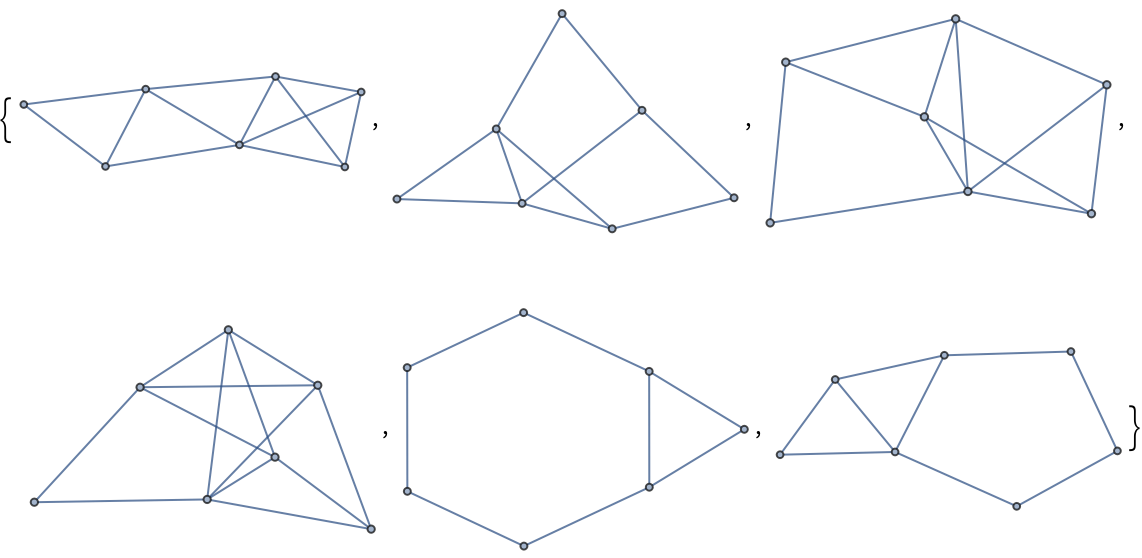 |
| In[227]:= |
The specified graph from the Graph Atlas:
| In[228]:= |
| Out[228]= | 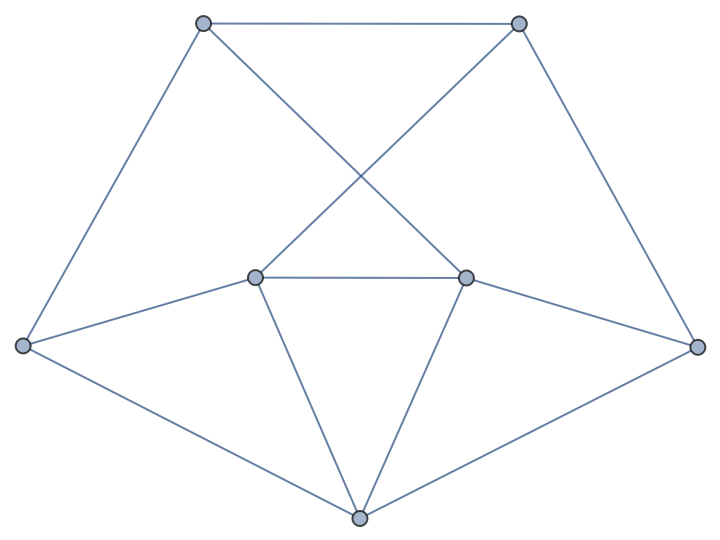 |
| In[229]:= |
A Zachary’s Karate Club graph:
| In[230]:= |
| Out[230]= | 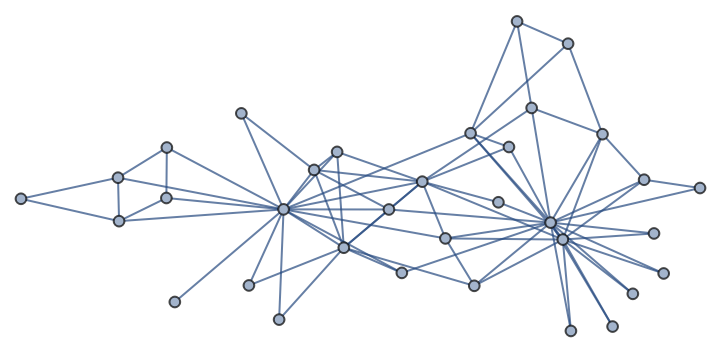 |
The same graph in the Wolfram Language (but note that NetworkX version is kept on the Python side):
| In[231]:= |
| Out[231]= | 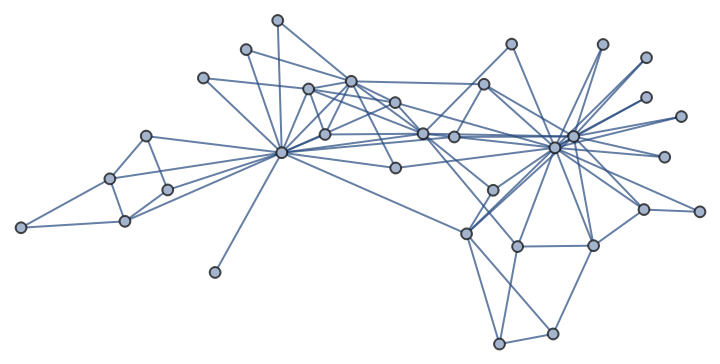 |
Create a series of random Spectral Graph Forge (SGF) graphs resembling the global properties of the previous graph:
| In[232]:= |
| Out[232]= | 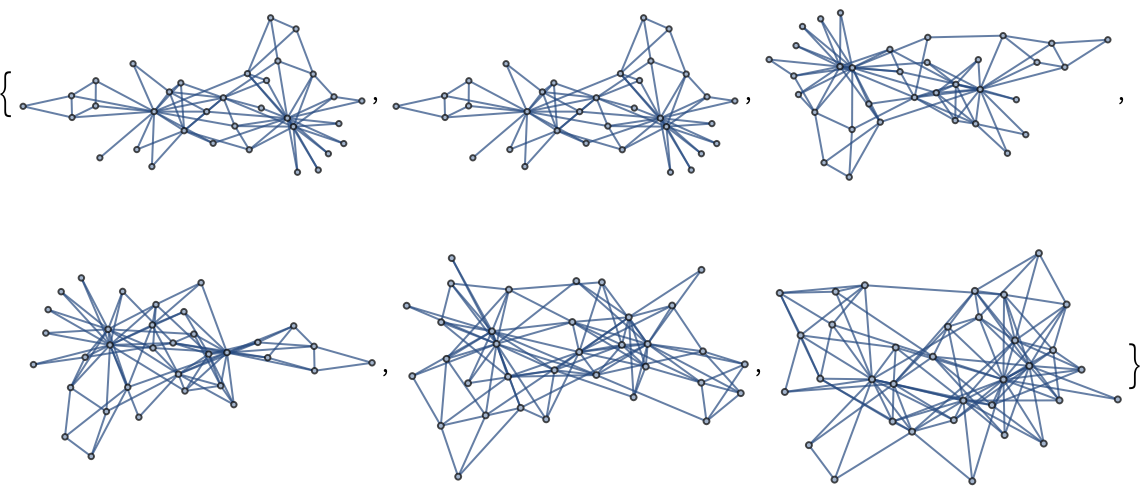 |
| In[233]:= |
A bipartite random graph with the first n nodes in the first bipartite set and the remaining m nodes in the second:
| In[234]:= | ![n = 5; m = 10;
nx = ResourceFunction["NetworkXObject"][];
nx["RandomGraph"[n, m, .75, "Seed" -> 1]] // Normal](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/34479c91e8a09362.png) |
| Out[234]= | 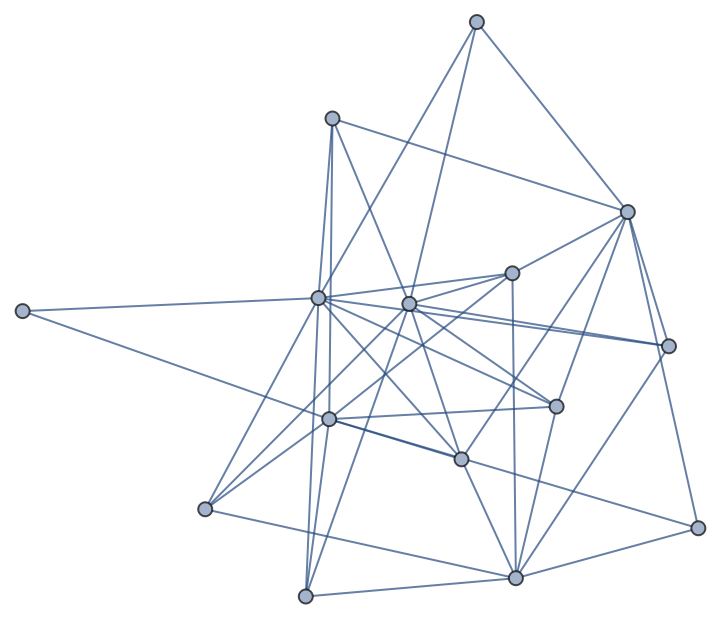 |
Show the bipartite partition of the graph:
| In[235]:= |
| Out[235]= | 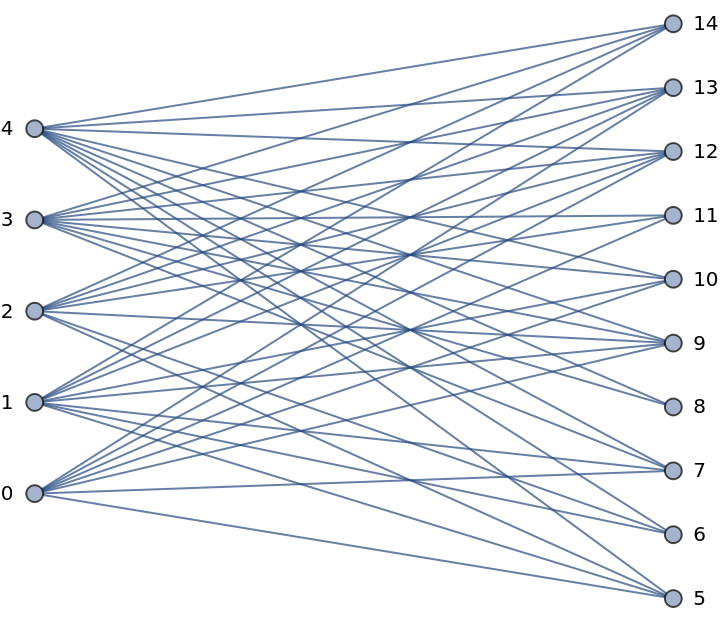 |
| In[236]:= |
A random partition graph:
| In[237]:= |
| Out[237]= | 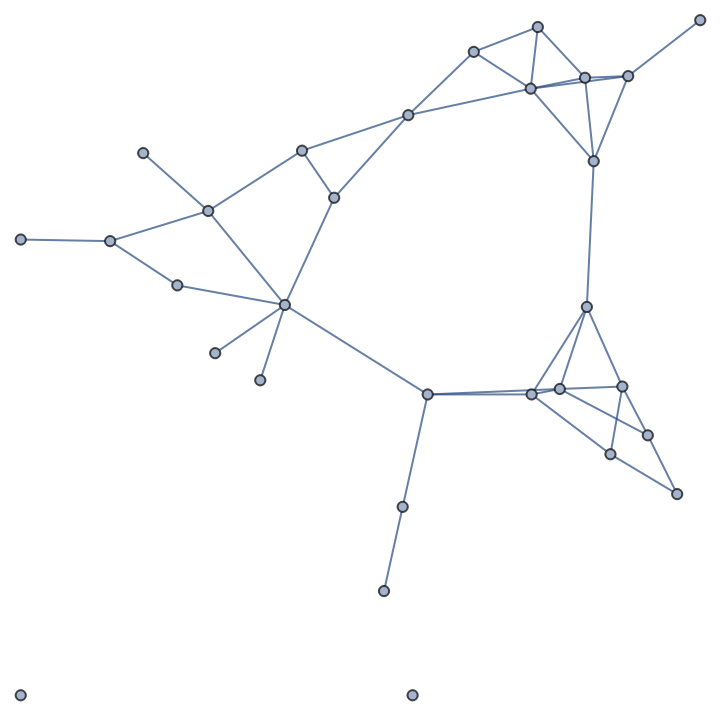 |
| In[238]:= |
| Out[238]= |
| In[239]:= |
A Gaussian random partition graph:
| In[240]:= |
| Out[240]= | 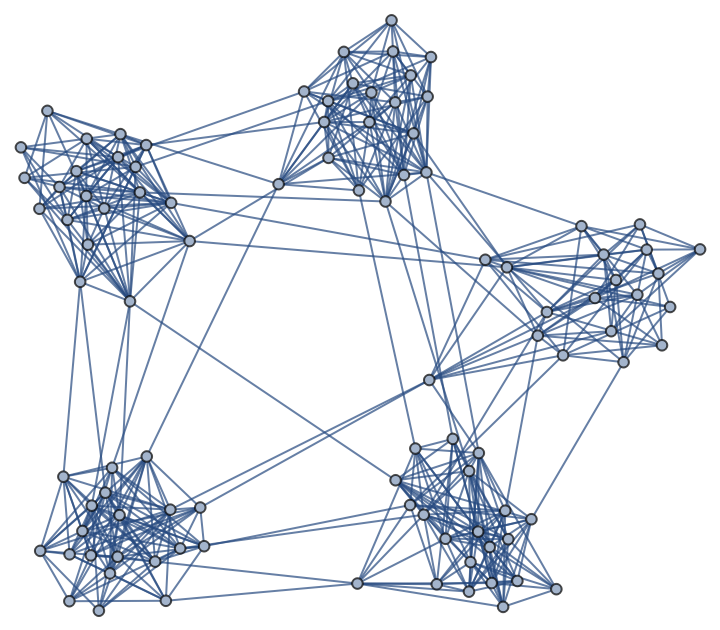 |
| In[241]:= |
| Out[241]= |  |
Highlight the partitions:
| In[242]:= |
| Out[242]= | 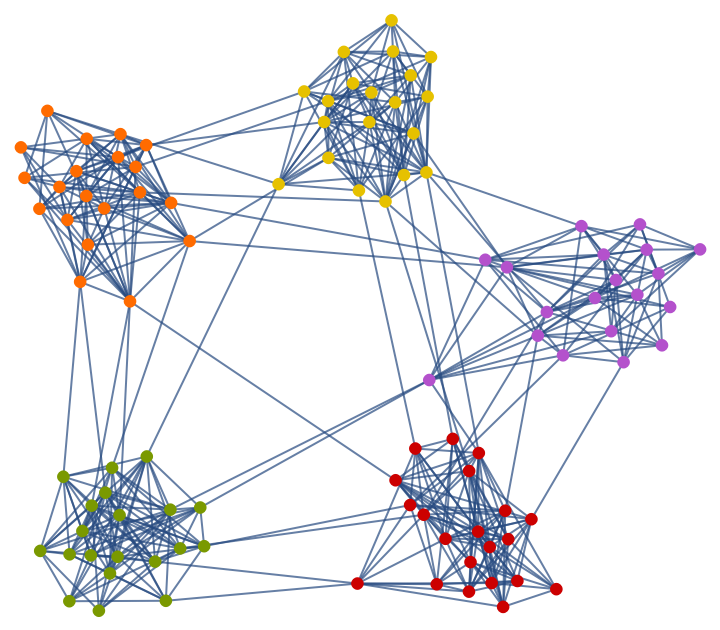 |
Explore functional interface to graph methods and assorted graph utilities:
| In[243]:= |
| Out[243]= |  |
| In[244]:= |
Add a star to a graph, with the first node in the middle of the star and the other nodes connected to it:
| In[245]:= | ![nx = ResourceFunction["NetworkXObject"][];
g = nx["Graph"[]];
nx["AddStar"[g, {0, 1, 2, 3}]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/30e26c55383d5b1e.png) |
| In[246]:= |
| Out[246]= | 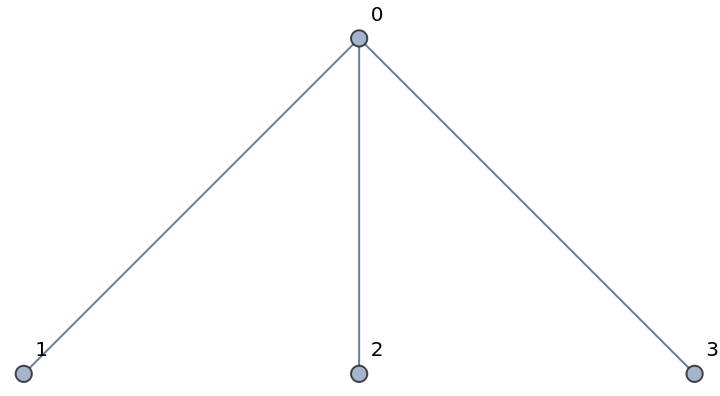 |
Add another star, with weighted edges:
| In[247]:= |
| In[248]:= |
| Out[248]= | 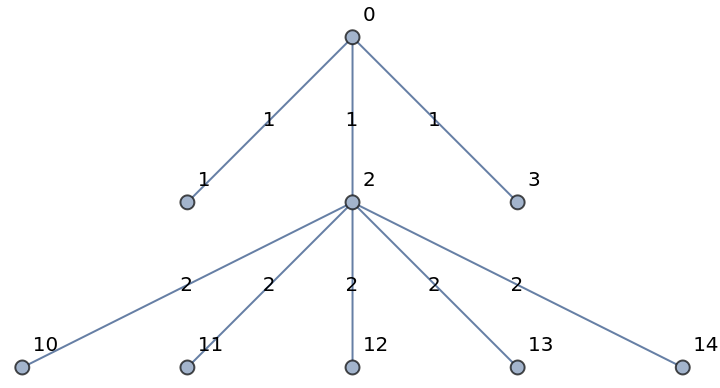 |
| In[249]:= |
Explore available graph-theoretic algorithms:
| In[250]:= |
| Out[250]= | 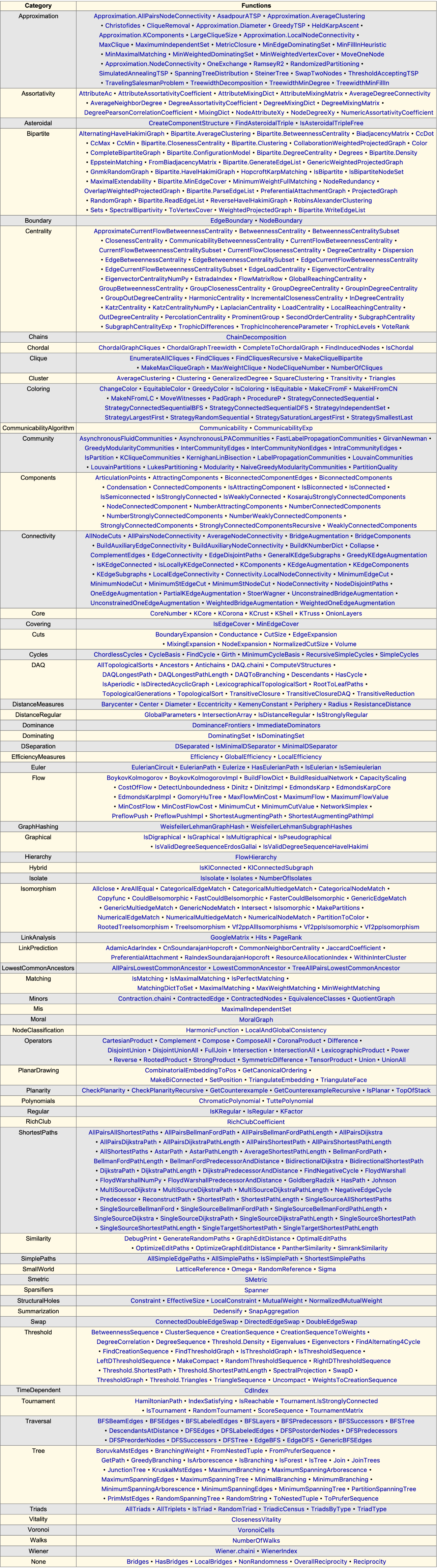 |
| In[251]:= |
Create a random bipartite graph with n nodes in the first bipartite set and compute the biadjacency matrix of the graph:
| In[252]:= | ![nx = ResourceFunction["NetworkXObject"][];
n = 5;
g = nx["RandomGraph"[n, 10, .75, "Seed" -> 1]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/4aec1480cb3ea0ab.png) |
| Out[252]= |
| In[253]:= |
| Out[253]= |  |
| In[254]:= |
| Out[254]= |  |
Despite being smaller, the biadjacency matrix fully describes the graph:
| In[255]:= |
| Out[255]= | 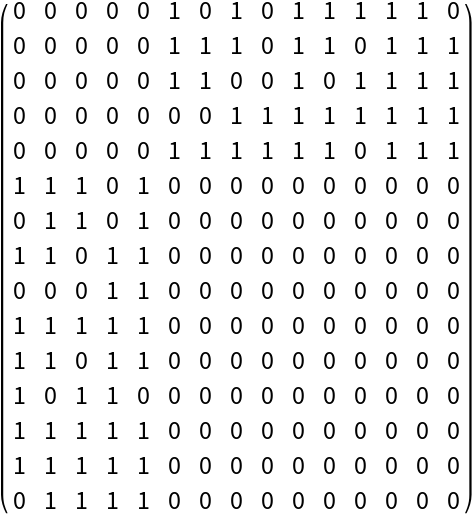 |
| In[256]:= |
| Out[256]= |
| In[257]:= |
Compute the clustering coefficient for nodes of a complete graph:
| In[258]:= |
| Out[258]= | 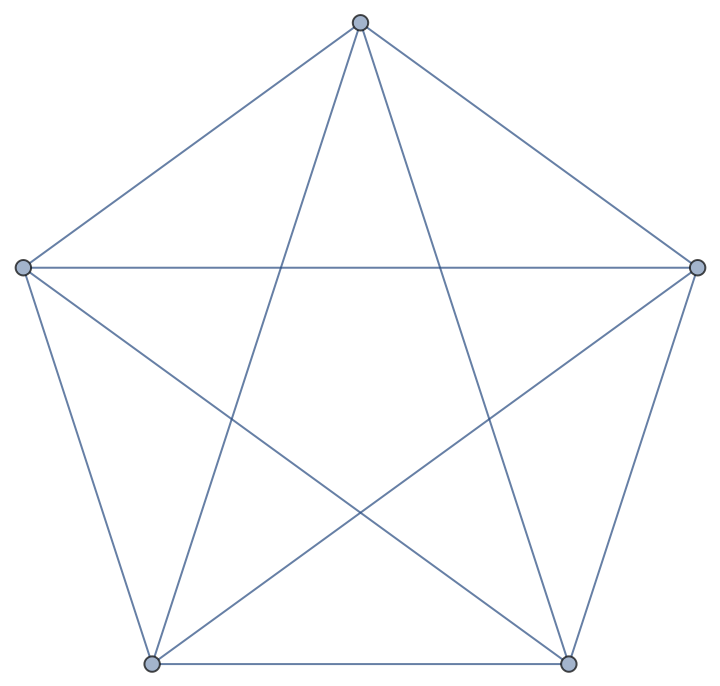 |
| In[259]:= |
| Out[259]= |
The clustering coefficient for the specified nodes:
| In[260]:= |
| Out[260]= |
For a single node:
| In[261]:= |
| Out[261]= |
| In[262]:= |
Compute the average clustering coefficient for nodes of a random Erdős-Rényi (binomial) graph:
| In[263]:= |
| Out[263]= | 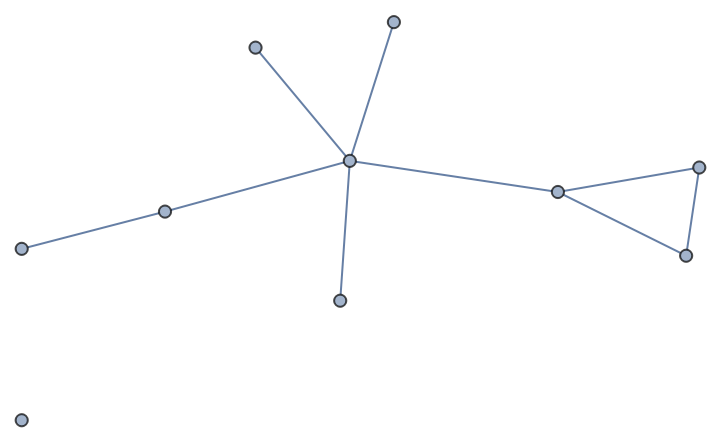 |
| In[264]:= |
| Out[264]= |
| In[265]:= |
Find the dominating set of a random geometric graph:
| In[266]:= |
| Out[266]= | 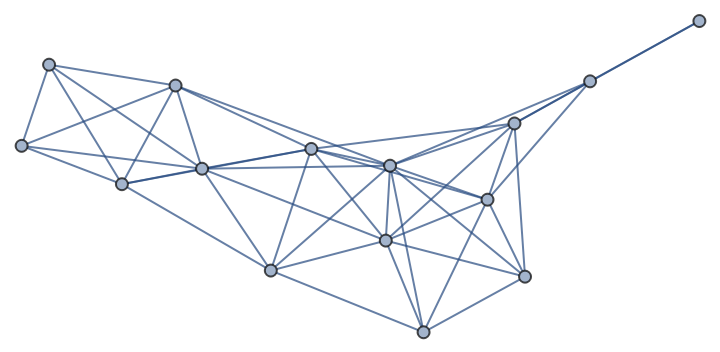 |
| In[267]:= |
| Out[267]= |
Confirm that the set of vertices form a dominating set:
| In[268]:= |
| Out[268]= | 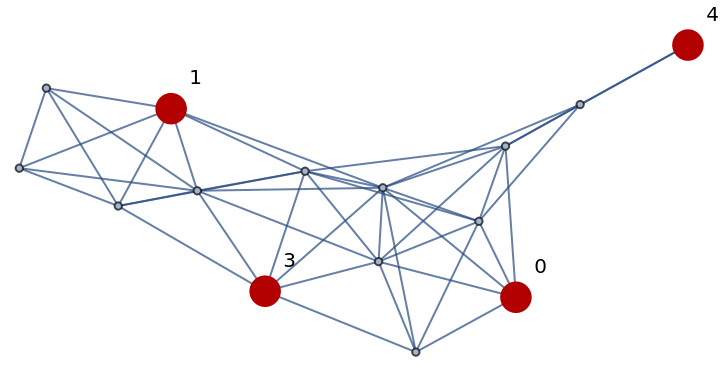 |
| In[269]:= |
| Out[269]= |
| In[270]:= |
Define a hidden Markov model (HMM) graph with five states and observation nodes:
| In[271]:= |
| Out[271]= |
| In[272]:= |
Check if states/observation nodes before "S3" are d-separated from the states/observations after "S3":
| In[273]:= |
| In[274]:= |
| In[275]:= |
| In[276]:= |
| Out[276]= |
| In[277]:= |
| Out[277]= | 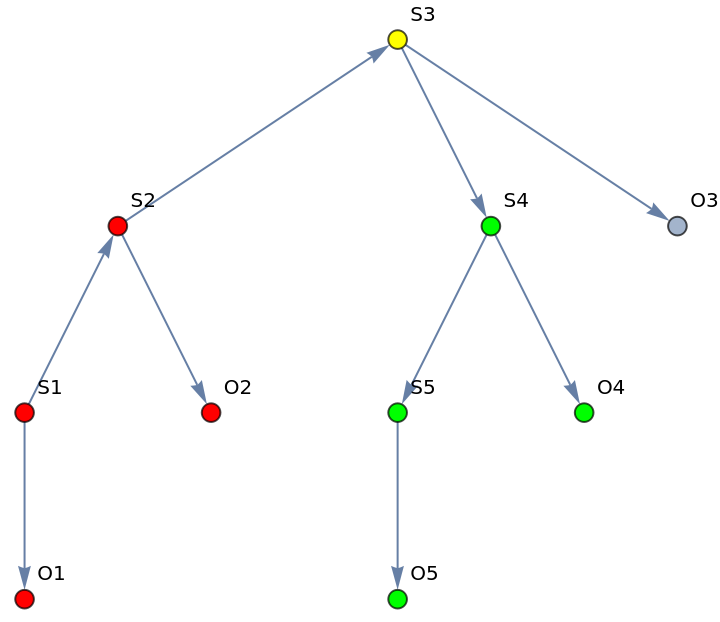 |
| In[278]:= |
Create an undirected moralized graph of given a directed graph:
| In[279]:= |
| Out[279]= |
| In[280]:= |
| Out[280]= |
Display the graphs:
| In[281]:= |
| Out[281]= | 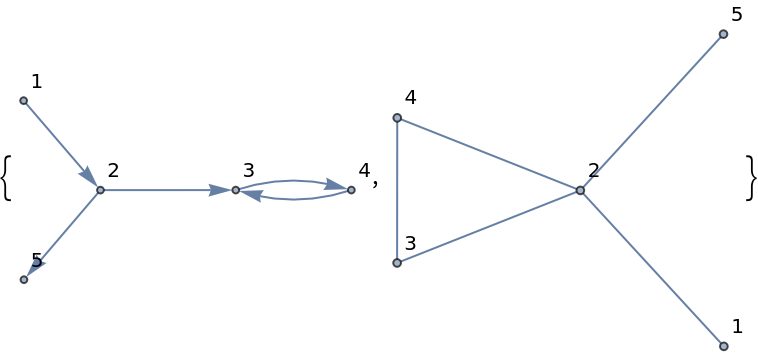 |
The added edge between unconnected parents that have a common child:
| In[282]:= |
| Out[282]= |
| In[283]:= |
Functions for drawing graphs include interfaces to Python packages PyGraphviz, PyDot, PyLab:
| In[284]:= |
| Out[284]= | 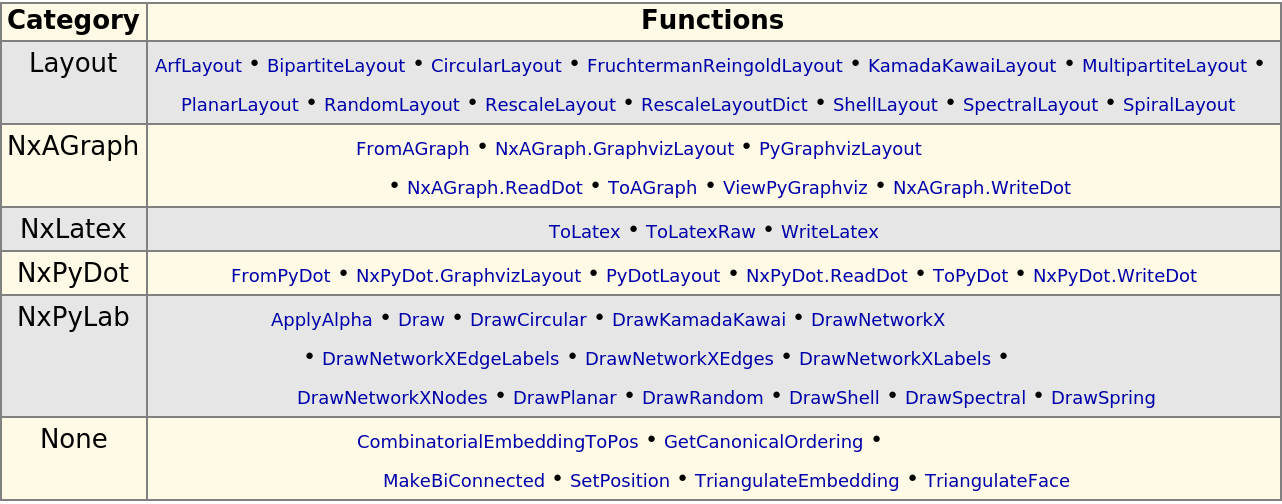 |
| In[285]:= |
Create a Matplotlib object to draw a graph on the Python side:
| In[286]:= |
| Out[286]= |
Prepare a subplot object to plot a graph:
| In[287]:= |
| Out[287]= |
Create and plot the graph in Python:
| In[288]:= |
| Out[288]= |
| In[289]:= |
| Out[289]= |
| In[290]:= |
Import the graph image:
| In[291]:= |
| Out[291]= | 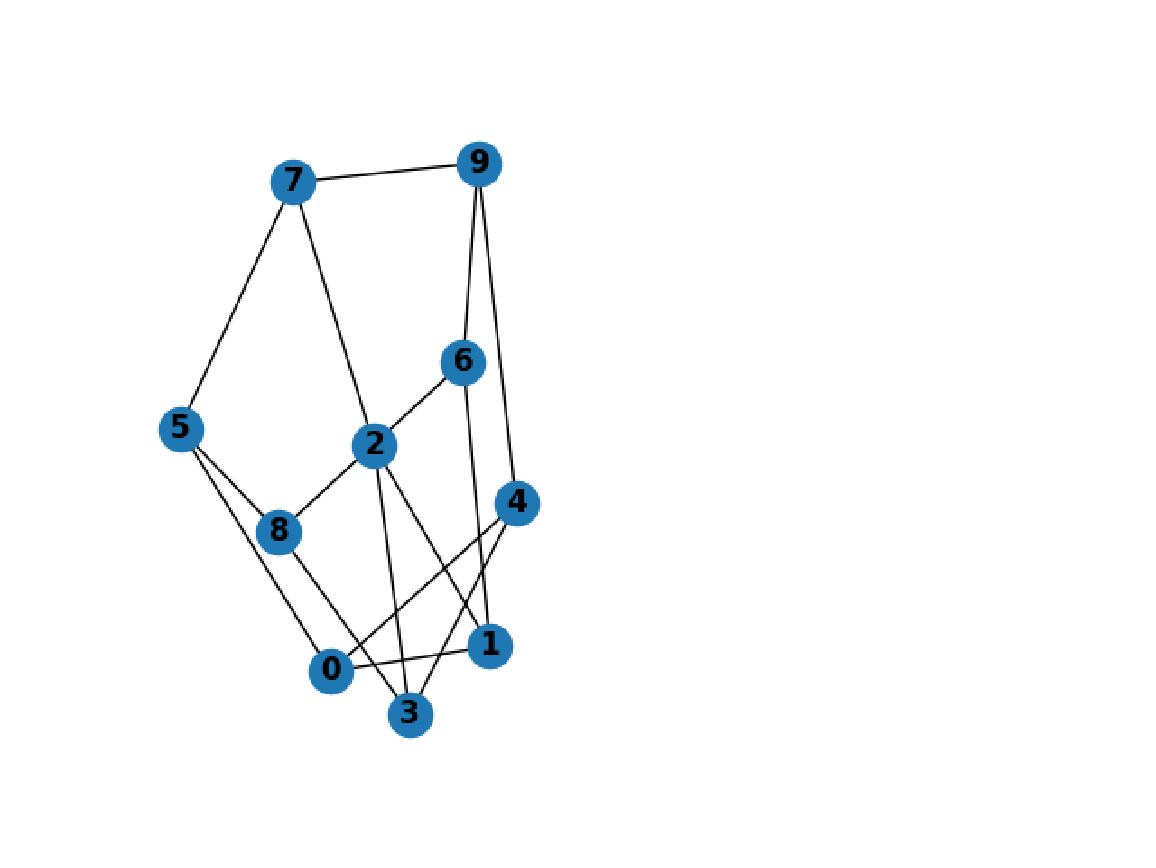 |
Prepare another subplot and draw the graph with shell layout:
| In[292]:= |
| Out[292]= |
| In[293]:= | ![nx["DrawShell"[g, "nlist" -> {Range[5, 9], Range[0, 4]}, "WithLabels" -> True, "FontWeight" -> "bold"]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/7e4a03a71f123234.png) |
Import the combined image:
| In[294]:= |
| Out[294]= | 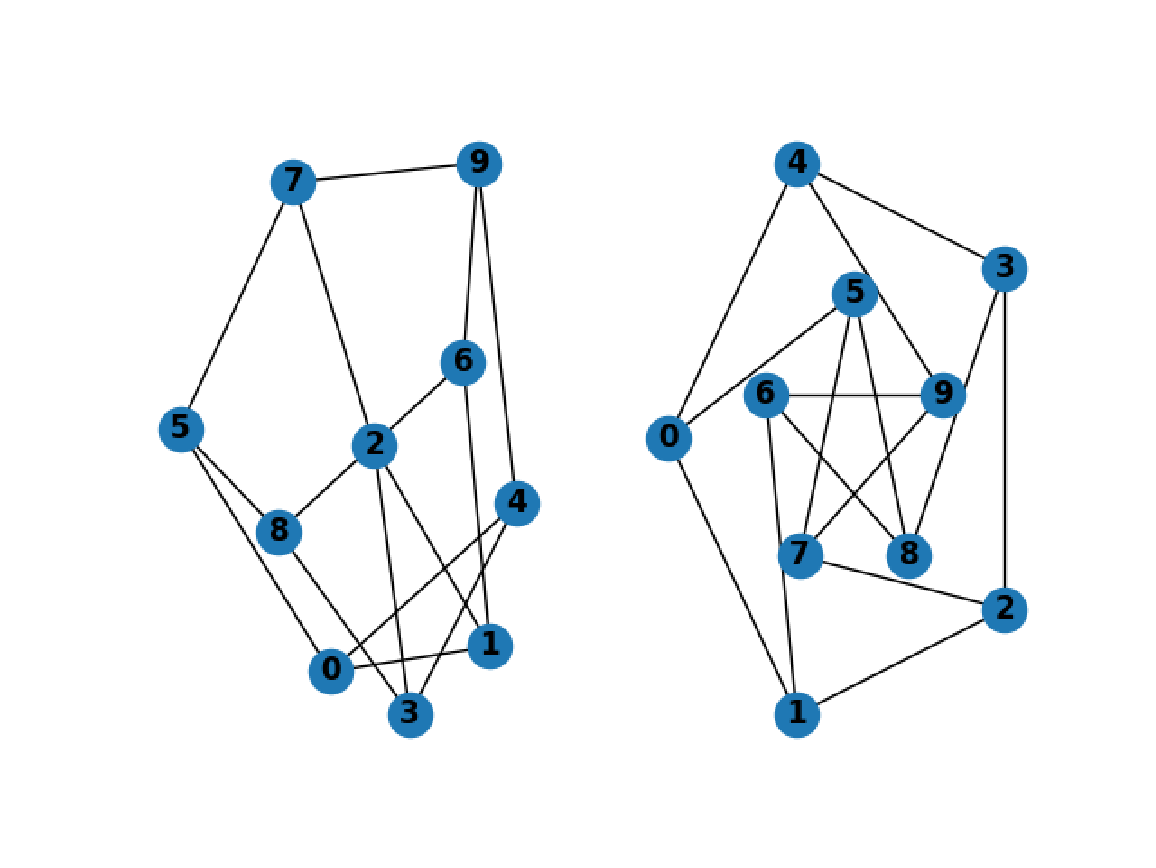 |
| In[295]:= |
Display a table graph-theoretical functions for linear algebra:
| In[296]:= |
| Out[296]= | 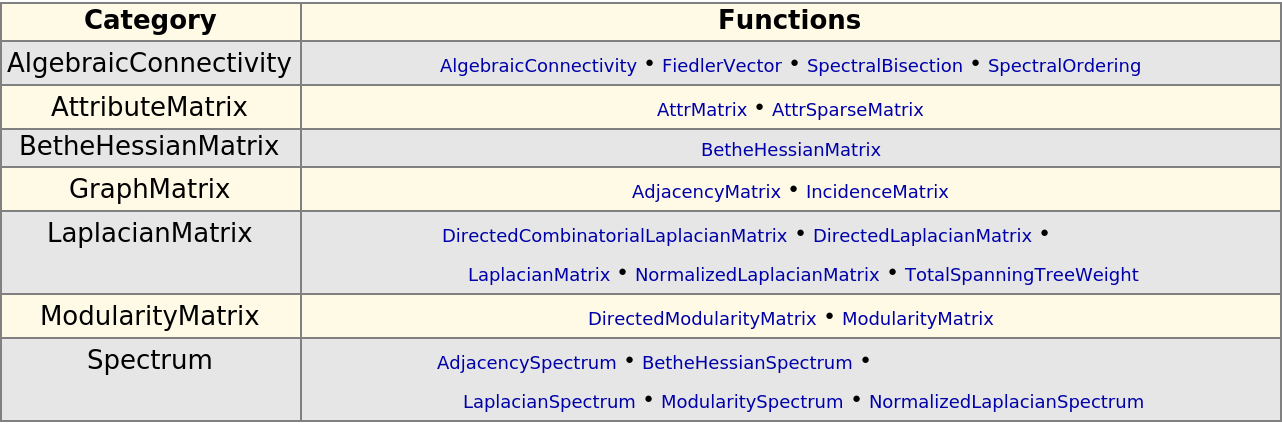 |
| In[297]:= |
Construct a simple graph with given degree sequence using the Havel-Hakimi algorithm:
| In[298]:= |
| Out[298]= |  |
The Bethe Hessian matrix of the graph:
| In[299]:= |
| Out[299]= |  |
| In[300]:= |
| Out[300]= | 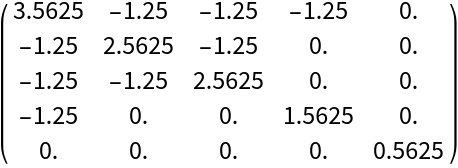 |
| In[301]:= |
Display a table of functions that import, export and process data in common graph formats:
| In[302]:= |
| Out[302]= | 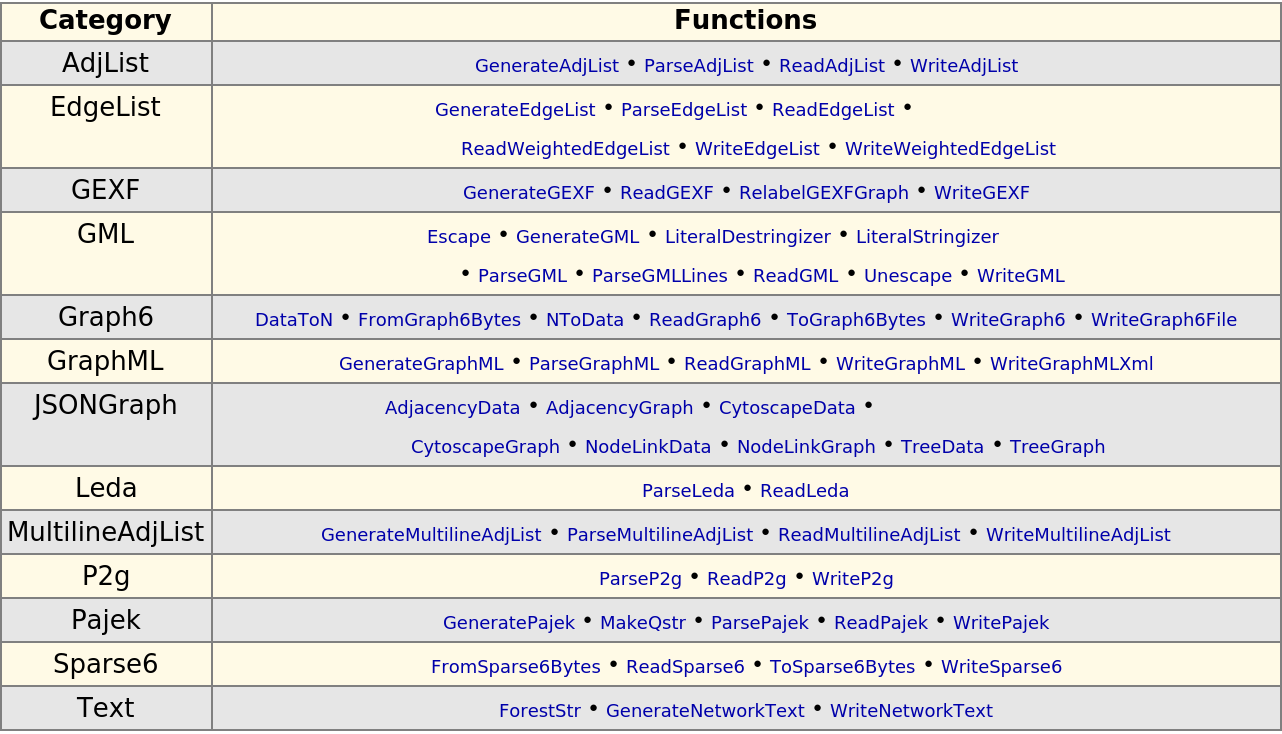 |
| In[303]:= |
Create a graph:
| In[304]:= |
| Out[341]= |  |
Export the graph in the Graph6 format:
| In[342]:= |
| In[343]:= |
Import the file in Python:
| In[344]:= |
| Out[344]= |
| In[345]:= |
| Out[345]= |  |
Import the file in the Wolfram Language:
| In[346]:= |
| Out[346]= | 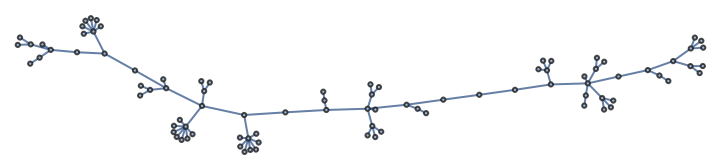 |
| In[347]:= |
Explore the available graph utilities:
| In[348]:= |
| Out[348]= | 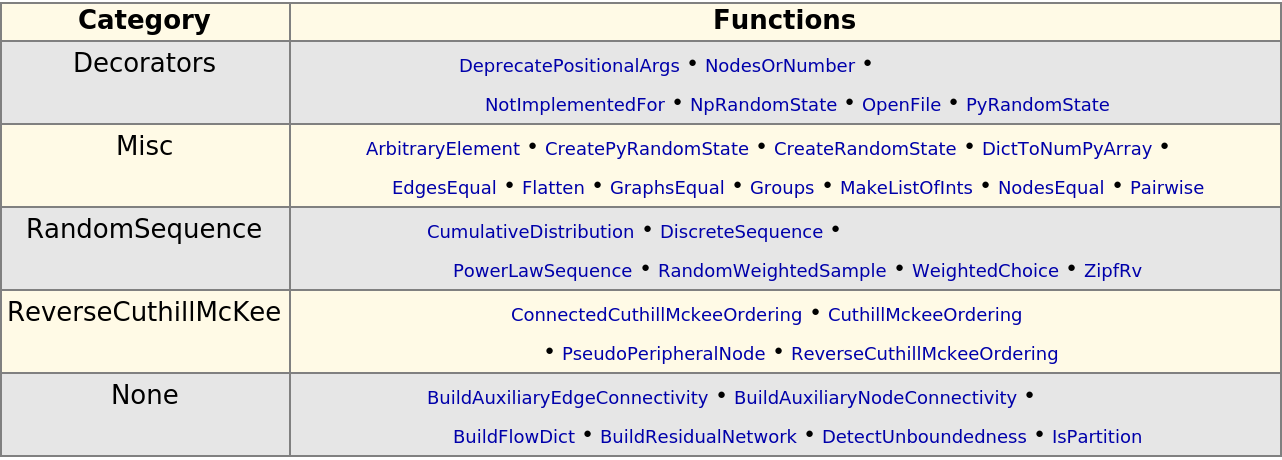 |
Utilities called by graph constructors that can also be called directly (and functions for converting back):
| In[349]:= |
| Out[349]= |  |
| In[350]:= |
Create an edge list using "Pairwise":
| In[351]:= |
| Out[64]= |  |
Partition creates an analogous list:
| In[352]:= |
| Out[352]= |
Create graphs from each to show the equivalence:
| In[353]:= |
| Out[353]= | 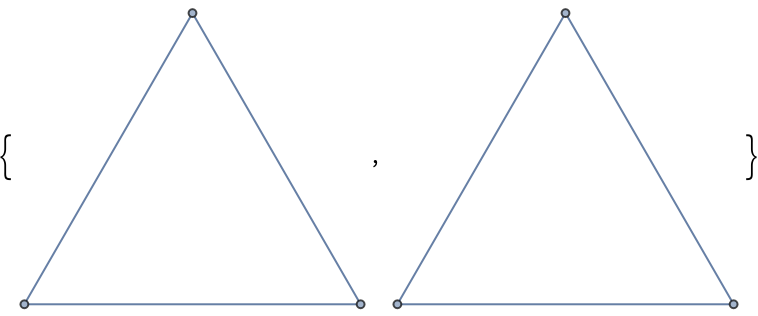 |
| In[354]:= |
Convert a graph to a SciPy sparse array:
| In[355]:= |
| Out[355]= |
| In[356]:= |
| Out[356]= |  |
Import the matrix:
| In[357]:= |
| Out[357]= |
| In[358]:= |
Control the type of returned object in graph constructors using the option CreateUsing:
| In[359]:= |
| Out[359]= |  |
| In[360]:= |
| Out[360]= | 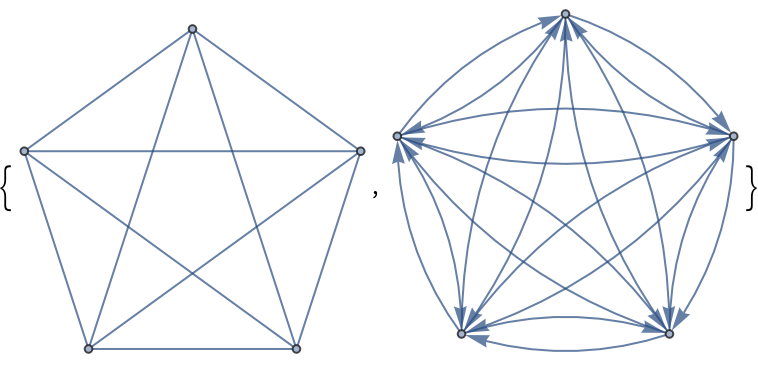 |
| In[361]:= |
Functions, such as "RelabelNodes", that have the option "Copy" allow you to modify the input graph object in-place:
| In[362]:= |
| Out[362]= |  |
By default, "RelabelNodes" returns a modified graph keeping the original intact:
| In[363]:= |
| Out[363]= |
Define the remapping:
| In[364]:= |
| Out[364]= |
| In[365]:= |
| Out[365]= |
Show the original and remapped vertex names:
| In[366]:= |
| Out[366]= |
Use "Copy"→False to modify the input graph in-place:
| In[367]:= |
| Out[367]= |
The graph g has changed:
| In[368]:= |
| Out[368]= | 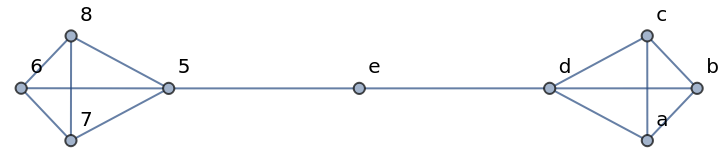 |
| In[369]:= |
The option "Seed" seeds the random number generator in functions that use randomness to generate, draw and compute properties or manipulate graphs.
Create a line graph:
| In[370]:= |
| Out[370]= |
Define a function that computes random coordinates of a graph, passing options to RandomLayout:
| In[371]:= |
Without a seed, executing the function twice produces different sets of random coordinates:
| In[372]:= |
| Out[372]= | 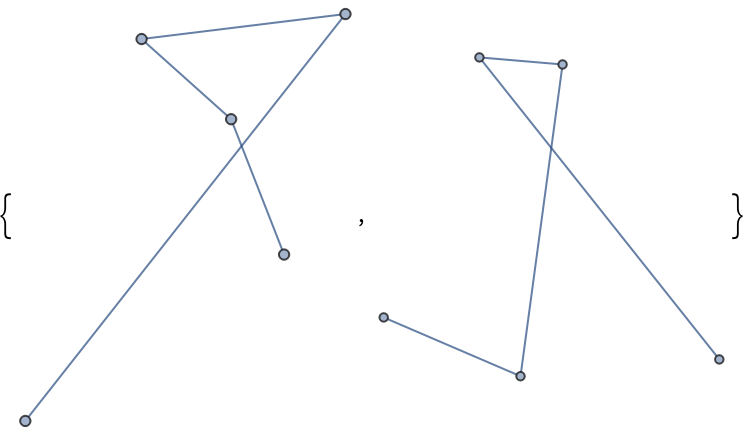 |
Use the option "Seed" to create a reproducible random layout:
| In[373]:= |
| Out[373]= | 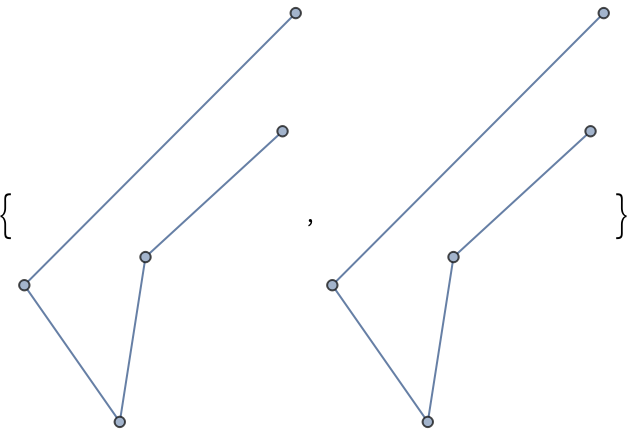 |
| In[374]:= |
Employ algorithms that might not yet be implemented in the Wolfram Language; for instance, find an approximate solution to the asymmetric traveling salesman problem using the algorithm developed by Asadpour et al.
Create a weighted directed graph:
| In[375]:= | ![nx = ResourceFunction["NetworkXObject"][];
CompleteGraph[3, DirectedEdges -> True, EdgeWeight -> {1 \[DirectedEdge] 2 -> 2, 2 \[DirectedEdge] 3 -> 2, 3 \[DirectedEdge] 1 -> 2, 1 \[DirectedEdge] 3 -> 1, 3 \[DirectedEdge] 2 -> 1, 2 \[DirectedEdge] 1 -> 1}, VertexLabels -> Automatic, EdgeLabels -> "EdgeWeight"]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/5b14a8e1bf1cdd4b.png) |
| Out[375]= | 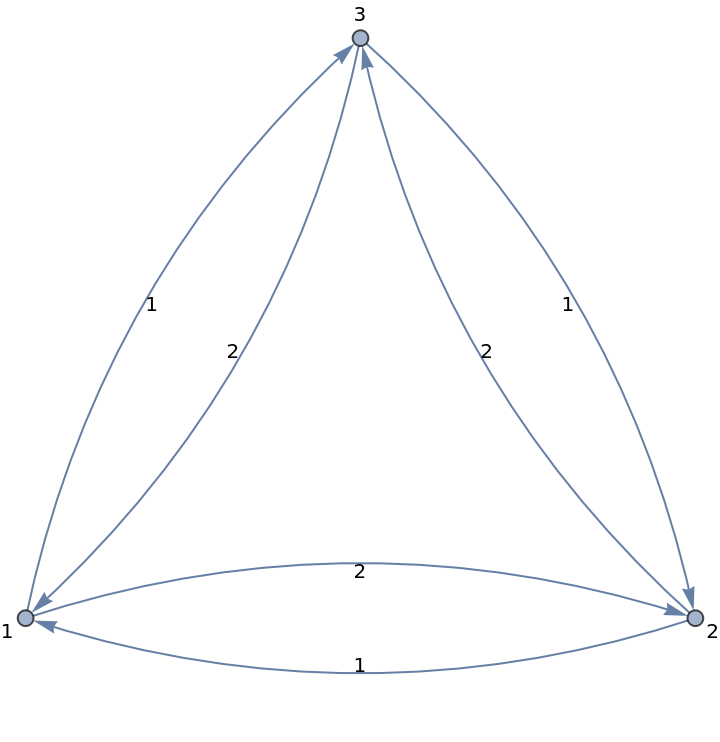 |
Import the graph in Python:
| In[376]:= |
| Out[376]= |
Find the shortest cycle starting and ending at vertex 1:
| In[377]:= |
| Out[377]= |
| In[378]:= |
Create a graph using a constructor that might not yet be implemented in the Wolfram Language, such as a directed graph constructor for the growing network with redirection (the GNR model):
| In[379]:= |
| Out[379]= | 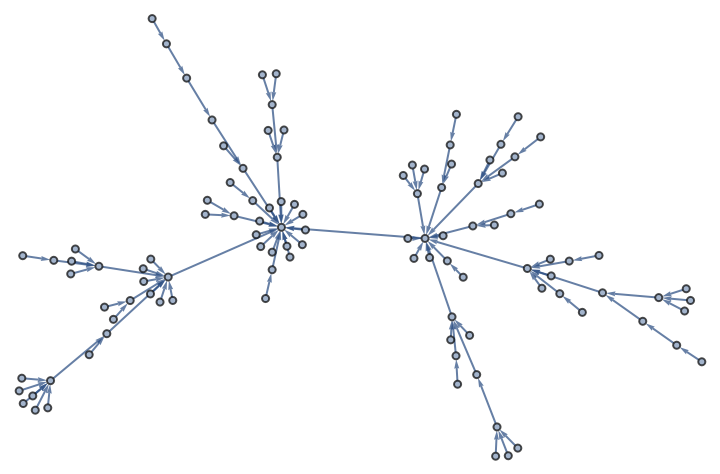 |
| In[380]:= |
Visualize data and draw conclusions from a pandas DataFrame object.
Create a small group of participants:
| In[381]:= | ![n = 20;
SeedRandom[10];
names = ResourceFunction["RandomPetName"][n]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/309552405a42729a.png) |
| Out[380]= |
Simulate a table exported from a JIRA bug database:
| In[382]:= |
| In[383]:= |
| Out[383]= |
Create a pandas "DataFrame" object for the database:
| In[384]:= | ![session = StartExternalSession["Python"];
pd = ResourceFunction["PandasObject"][session];
df = pd["DataFrame"[db]]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/6cc92cd204cd9416.png) |
| Out[384]= |
| In[385]:= |
| Out[385]= | 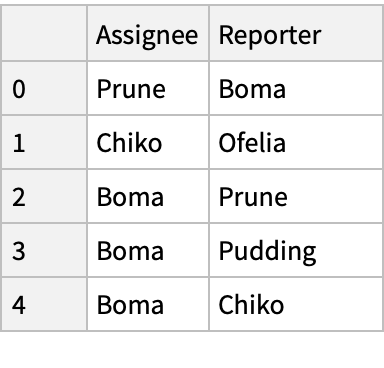 |
Construct a graph of the database:
| In[386]:= |
| Out[386]= |
| In[387]:= |
| Out[387]= |
Team members Boma and Pudding are apparently in the center of action:
| In[388]:= |
| Out[388]= | 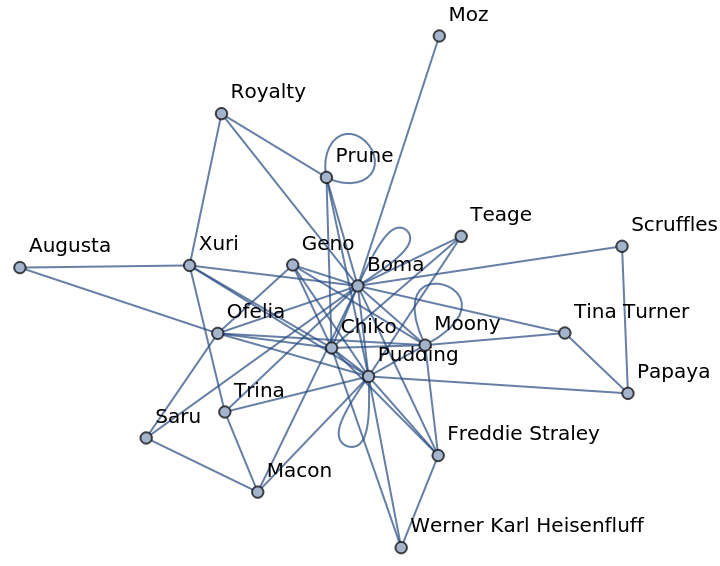 |
Extract all connections represented by edges of the graph:
| In[389]:= |
| Out[389]= |  |
Create a table of participants sorted by activity:
| In[390]:= |
| In[391]:= |
| Out[391]= |  |
| In[392]:= |
NetworkXObject[…] gives the same result as the resource function PythonObject with a special configuration:
| In[393]:= |
| Out[393]= |
| In[394]:= |
| Out[394]= |
| In[395]:= |
Get information on a NetworkX object:
| In[396]:= |
| Out[396]= |
| In[397]:= |
| Out[397]= |
Open the user guide in your default web browser:
| In[398]:= |
| In[399]:= |
Some of the functions and classes available in the NetworkX module:
| In[400]:= |
| Out[400]= |
| In[401]:= |
| Out[401]= |
| In[402]:= |
Information on a function:
| In[403]:= |
| Out[403]= |  |
Web documentation of a function:
| In[404]:= |

| In[405]:= |
Functions similar to the given one; click the function name to go to the web documentation:
| In[406]:= |
| Out[406]= |
| In[407]:= |
| Out[407]= | 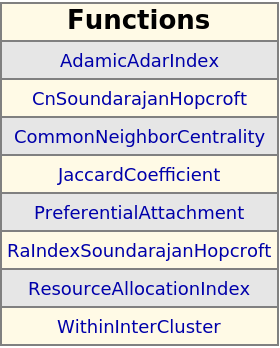 |
| In[408]:= |
Aliases defined for a function or class:
| In[409]:= |
| Out[409]= |
| In[410]:= |
| Out[410]= |
| In[411]:= |
NetworkX Graph objects are analogous to Graph, but keep the objects on the Python side:
| In[412]:= |
| Out[412]= | 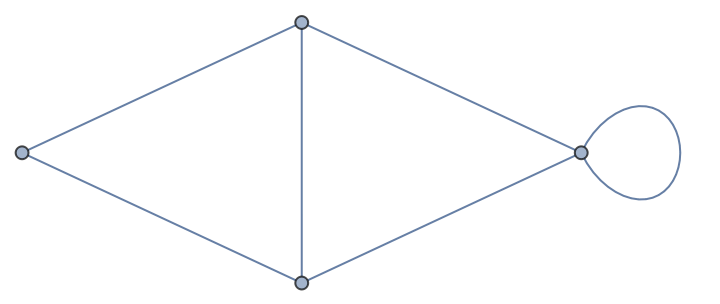 |
| In[413]:= |
| Out[413]= |
Similarly, many graph properties and functions closely correspond to Wolfram Language functions:
| In[414]:= |
| Out[414]= |
| In[415]:= |
| Out[415]= |
| In[416]:= |
Functions of the same name might be defined in different modules:
| In[417]:= |
| Out[417]= |
| In[418]:= |
| In[419]:= |
| Out[419]= |  |
Functions defined for both general and bipartite graphs:
| In[420]:= |
| Out[420]= |  |
| In[421]:= |
Define a bipartite graph in the Wolfram Language and NetworkX:
| In[422]:= |
| Out[422]= | 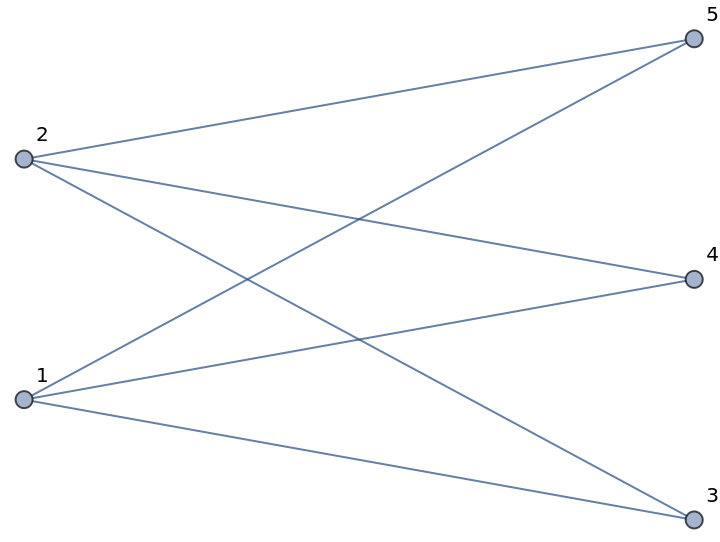 |
| In[423]:= |
| Out[423]= |
The general function "ClosenessCentrality" function can be accessed by an unqualified name, returning the same values as the corresponding Wolfram Language function ClosenessCentrality:
| In[424]:= |
| Out[424]= |
| In[425]:= |
| Out[425]= |
Use a qualified name to access the bipartite version of the function:
| In[426]:= |
| Out[426]= |
| In[427]:= |
NetworkX uses zero-based indexes, while the Wolfram Language Graph uses one-based node indexes:
| In[428]:= |
| Out[428]= |
| In[429]:= |
| Out[429]= |
See the vertex indexing from NetworkX:
| In[430]:= |
| Out[430]= |
Compare with the default indexing in Graph:
| In[431]:= |
| Out[431]= |
Use "RelabelNodes" to index the nodes differently:
| In[432]:= |
| Out[432]= |
Now the indices are identical:
| In[433]:= |
| Out[433]= |
| In[434]:= |
When constructing graphs from Graph, NetworkXObject may not pass all annotations:
| In[435]:= | ![g = PathGraph[Range[3], AnnotationRules -> {1 -> {VertexLabels -> "hello"}, 2 -> {VertexLabels -> "there"}}]](https://www.wolframcloud.com/obj/resourcesystem/images/15d/15de1605-6c74-4421-8560-82793e1100aa/47e5f68d59513526.png) |
| Out[435]= |
| In[436]:= |
| Out[436]= |
Use "SetNodeAttributes" to pass the annotations:
| In[437]:= |
| Out[437]= |
| In[438]:= |
| In[439]:= |
| Out[439]= |
| In[440]:= |
NetworkXObject choses the type of edges in graph constructors based on the graph type, effectively discarding the DirectedEdge and UndirectedEdge wrappers:
| In[441]:= |
| Out[441]= |
| In[442]:= |
| Out[442]= |
Similarly, the wrappers are discarded in all other functions:
| In[443]:= |
| Out[443]= |
| In[444]:= |
An attempt to call a function by an unqualified name might fail if the function is defined in multiple contexts:
| In[445]:= |
| Out[445]= |  |
Use a qualified name to call the bipartite version of the function:
| In[446]:= |
| Out[446]= |
| In[447]:= |
| Out[447]= | 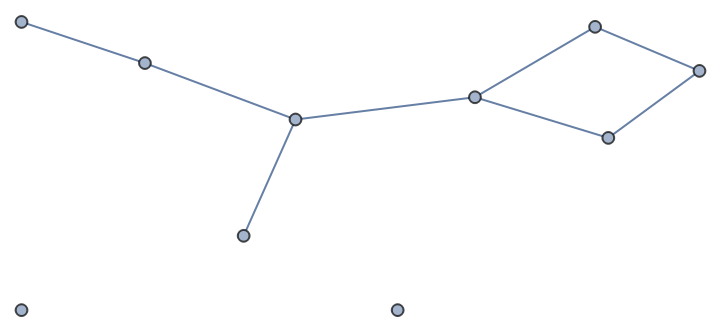 |
Use an unqualified name to call the general function:
| In[448]:= |
| Out[448]= |
| In[449]:= |
| Out[449]= |  |
| In[450]:= |
Wolfram Language 12.3 (May 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License