Details
Selected "
SNP" entities or dbSNP reference SNP numbers can be used.
ALFAFrequencyData is based on the
Allele Frequency Aggregator (ALFA) project. Allele frequency data from 12 diverse populations, including European, African, Asian, Latin American, and others, are provided.
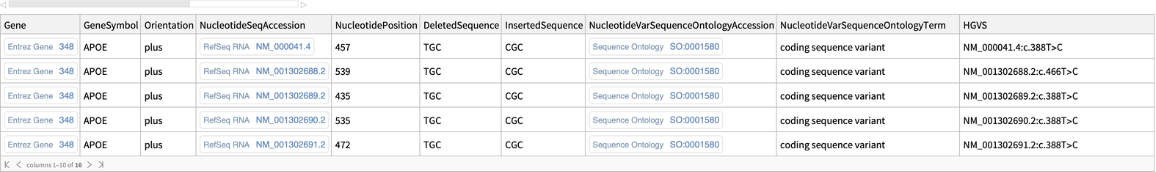
The "VariantDetails" content is a
Dataset containing known variant placements on genomic sequences with the following properties for each SNP:
| "Gene" | associated ID |
| "GeneSymbol" | associated gene symbol |
| "Orientation" | orientation of the genomic sequence |
| "NucleotideSeqAccession" | NCBI nucleotide sequence accession ID |
| "NucleotidePosition" | position of the allele on the nucleotide sequence |
| "DeletedSequence" | sequence of deleted nucleotides or the codon |
| "InsertedSequence" | sequence of inserted nucleotides or the codon |
| "NucleotideVarSequenceOntologyAccession" | accession ID of the Sequence Ontology (SO) concept describing the nucleotide sequence variation |
| "NucleotideVarSequenceOntologyTerm" | name of the Sequence Ontology (SO) concept describing the nucleotide sequence variation |
| "HGVS" | Human Genome Variation Society (HGVS) notation |
| "ProteinSeqAccession" | NCBI protein sequence accession ID |
| "ProteinPosition" | position of the amino acid change on the protein sequence |
| "DeletedAminoAcid" | letter of the deleted amino acid |
| "InsertedAminoAcid" | letter of the inserted amino acid |
| "ProteinVarSequenceOntologyAccession" | accession ID of the Sequence Ontology (SO) concept describing the protein sequence variation |
| "ProteinVarSequenceOntologyTerm" | name of the Sequence Ontology (SO) concept describing the protein sequence variation |
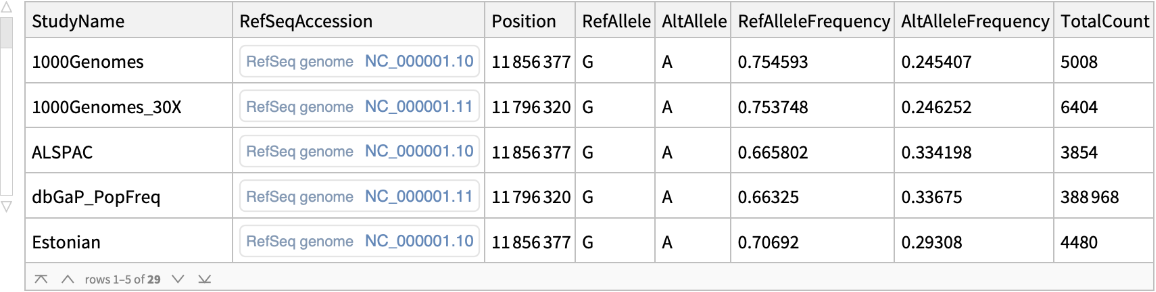
The "FrequencyData" content is a
Dataset of the reference and alternate allele frequencies reported by various studies:
| "StudyName" | name of the study |
| "RefSeqAccession" | NCBI refrence sequence accession ID |
| "Position" | position of the allele on the reference sequence |
| "RefAllele" | reference allele |
| "AltAllele" | alternate allele |
| "RefAlleleFrequency" | reported reference allele frequency |
| "AltAlleleFrequency" | reported alternate allele frequency |
| "TotalCount" | total sample size |
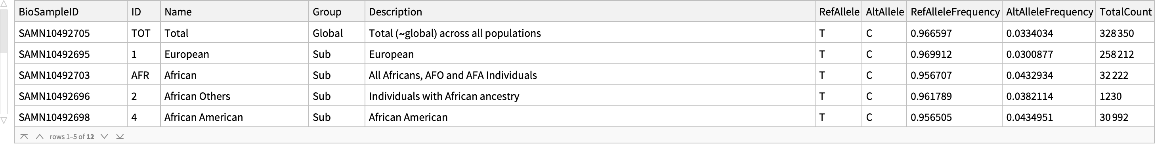
The "ALFAFrequencyData" content is a
Dataset of reference and alternate allele frequencies aggregated by population groups:
| "BioSampleID" | ID |
| "ID" | population ID |
| "Name" | population name |
| "Group" | population group |
| "Description" | population description |
| "RefAllele" | reference allele |
| "AltAllele" | alternate allele |
| "RefAlleleFrequency" | reported reference allele frequency |
| "AltAlleleFrequency" | reported alternate allele frequency |
| "TotalCount" | total sample size |
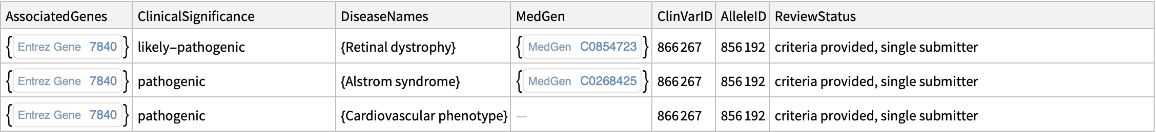
The "ClinicalSignificance" content is a
Dataset of clinical significance information from
ClinVar associated with the variations:
| "AssociatedGenes" | associated IDs |
| "ClinicalSignificance" | reported clinical significance |
| "DiseaseNames" | names of associated diseases |
| "MedGen" | associated MedGen concepts |
| "ClinVarID" | associated ClinVar ID |
| "AlleleID" | assigned allele ID reported in ClinVar |
| "ReviewStatus" | assigned review status |
ResourceFunction["NCBIGenomicSNPData"][snp] is equivalent to ResourceFunction["NCBIGenomicSNPData"][snp,"VariantDetails"].
![alt = StringReplacePart[ref, brca[1, "InsertedSequence"], {brca[1, "NucleotidePosition"], brca[1, "NucleotidePosition"] + StringLength[brca[1, "DeletedSequence"]] - 1}];](https://www.wolframcloud.com/obj/resourcesystem/images/6f2/6f2d5756-cc3b-42f3-932f-bc6d163c3291/1-2-0/3375ef0adfa271cb.png)

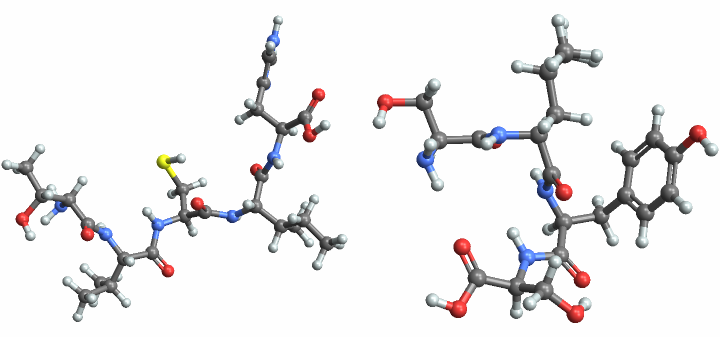
![refPep = BioSequenceTranslate[
BioSequence[
StringTake[
ref, {brca[1, "NucleotidePosition"], brca[1, "NucleotidePosition"] + 14}]]]](https://www.wolframcloud.com/obj/resourcesystem/images/6f2/6f2d5756-cc3b-42f3-932f-bc6d163c3291/1-2-0/1a485d8423e1455e.png)
![altPep = BioSequenceTranslate[
BioSequence[
StringTake[
alt, {brca[1, "NucleotidePosition"], brca[1, "NucleotidePosition"] + 14}]]]](https://www.wolframcloud.com/obj/resourcesystem/images/6f2/6f2d5756-cc3b-42f3-932f-bc6d163c3291/1-2-0/3b21c5a5ae482c36.png)