Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get the graph and additional information of a KEGG pathway
ResourceFunction["KEGGPathway"][organism,pathcode,property] get some property of the KEGG pathway associated to organism and pathcode. | |
ResourceFunction["KEGGPathway"][organism,pathcode,property,component] get some component of a property of the KEGG pathway associated to organism and pathcode. |
| KEGGPathway["Organisms"] | gets the list of organisms codes and names |
| KEGGPathway["Pathways"] | to get the list of pathways |
| KEGGPathway[organism,"Pathways"] | to get the list of pathways for an specific organism |
| "Information" | information of the KEGG pathway associated to an organism and pathcode |
| "Entries" | dataset of the entries in the KEGG pathway associated to an organism and pathcode |
| "Relations" | dataset of the relations between entries in the KEGG pathway associated to an organism and pathcode |
| "Reactions" | dataset of the reactions between entries in the KEGG pathway associated to an organism and pathcode |
| "Graph" | graph for any KEGG pathway associated to an organism and pathcode |
| "GraphRelation" | graph for a KEGG pathway associated to an organism and pathcode which only has relations between entries |
| "GraphReaction" | graph for a KEGG pathway associated to an organism and pathcode which only has reactions between entries |
| "GraphReactionAndRelation" | graph for a KEGG pathway associated to an organism and pathcode which has both reactions and relations |
| "EntryGraphName" | information for just the KEGG entry (gene,compound, reaction) named in EntryGraphName property of the "Entries" dataset |
| "Relation" | information for the relation |
| "Reaction" | information for the reaction |
| "Distance" | 0.017 | distance between the vertex and edge |
| "ArrowheadSize" | 0.013 | arrowhead size |
Get each organism name and its associated code:
| In[1]:= |
| Out[1]= | 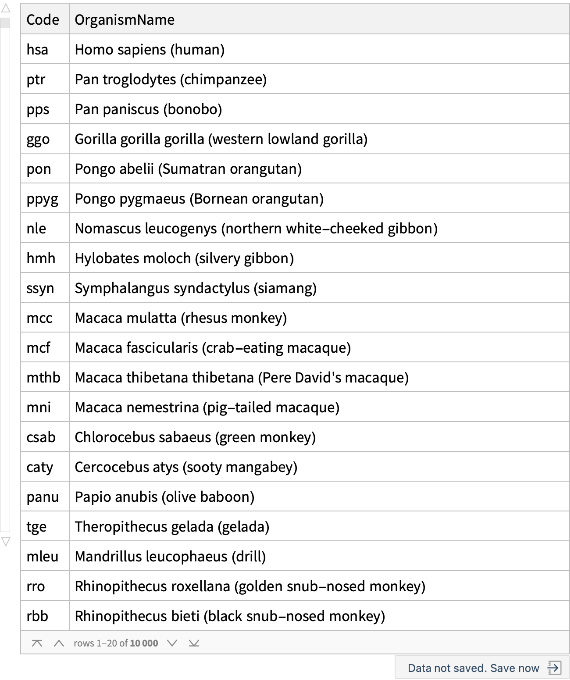 |
Get the long name of each pathway and its associated code:
| In[2]:= |
| Out[2]= | 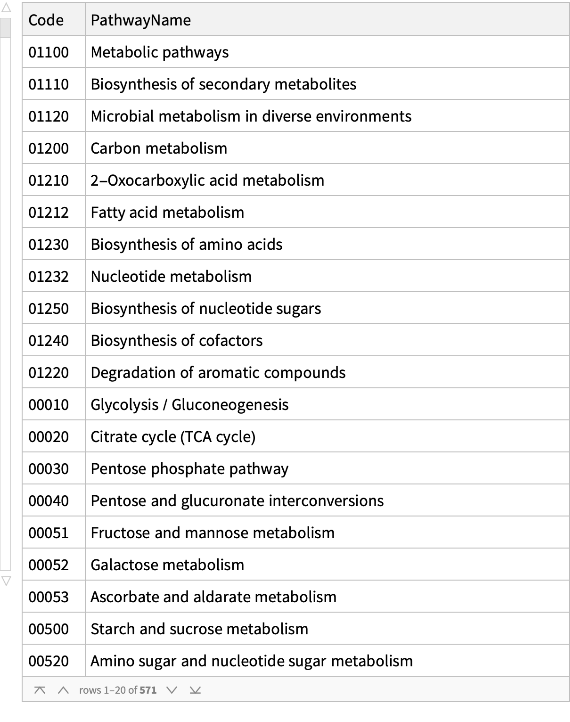 |
Get the pathways for a human:
| In[3]:= |
| Out[3]= | 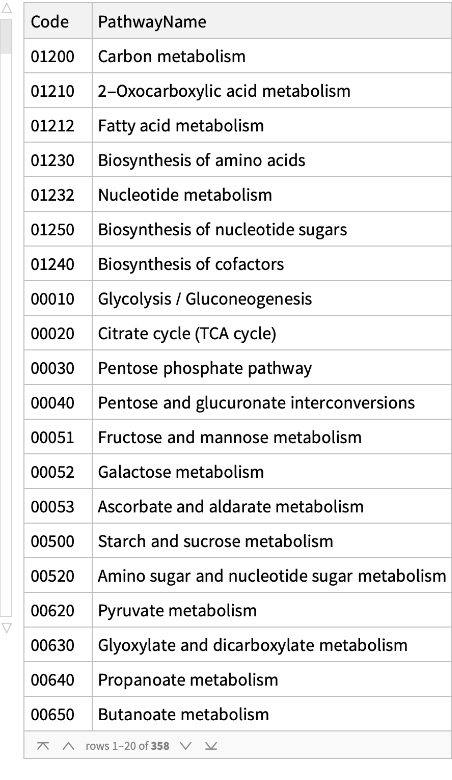 |
Get information about the pathway "00020" of the "hsa":
| In[4]:= |
| Out[4]= | 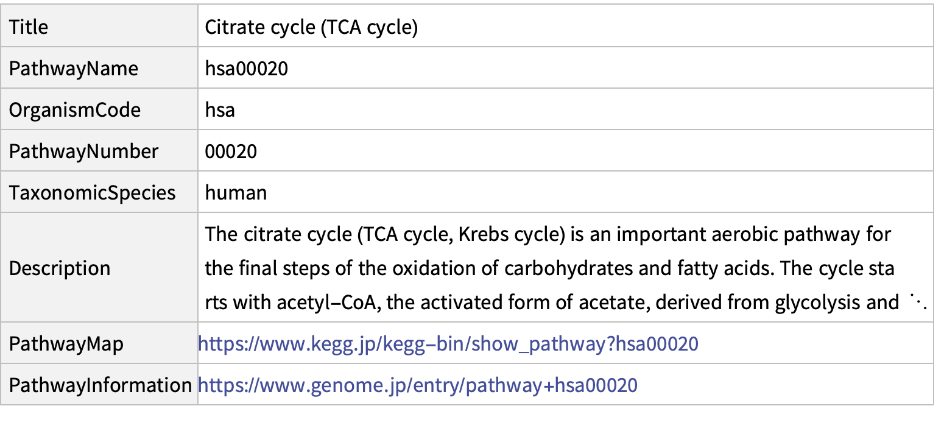 |
This is the alternate way to get same information as above:
| In[5]:= |
| Out[5]= | 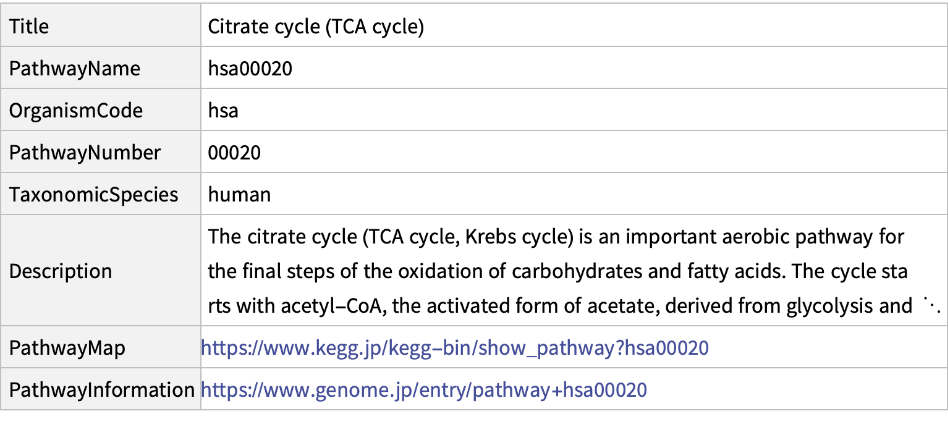 |
Get the entries of the citrate cycle (TCA cycle) of a human:
| In[6]:= |
| Out[14]= | 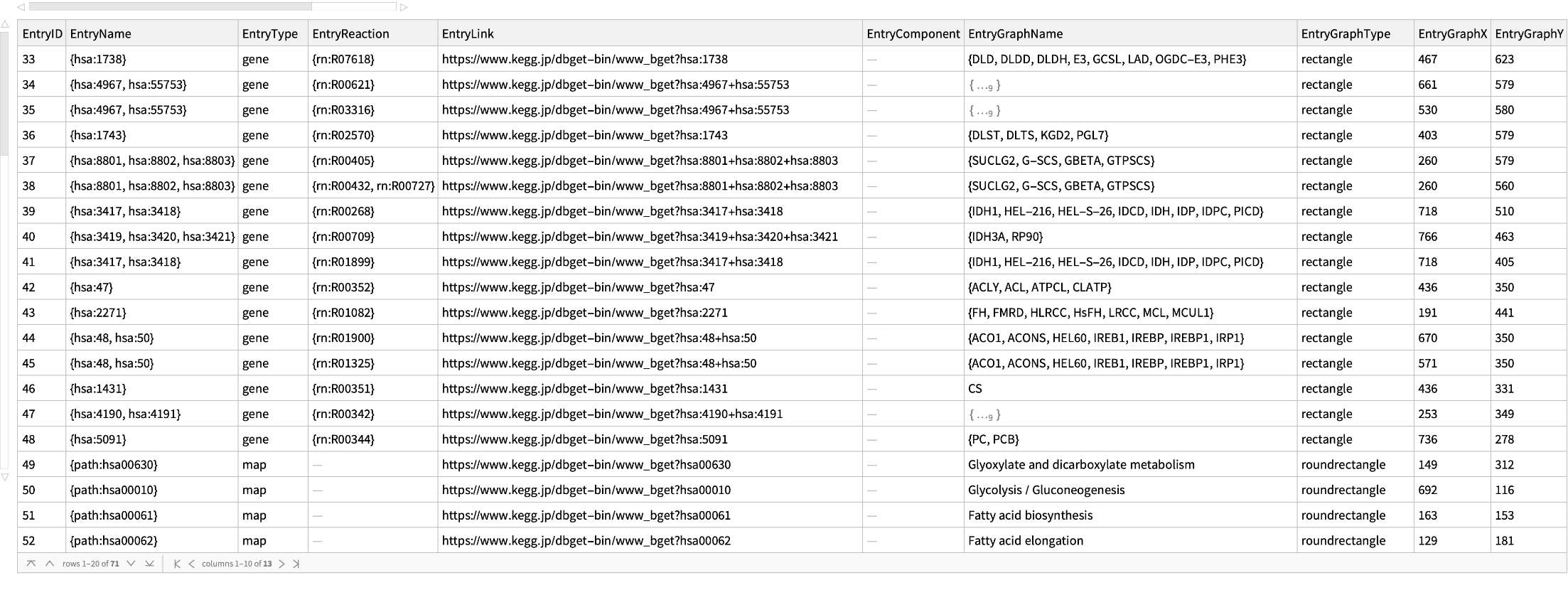 |
Get the relation entries of the citrate cycle (TCA cycle) of a human:
| In[15]:= |
| Out[15]= | 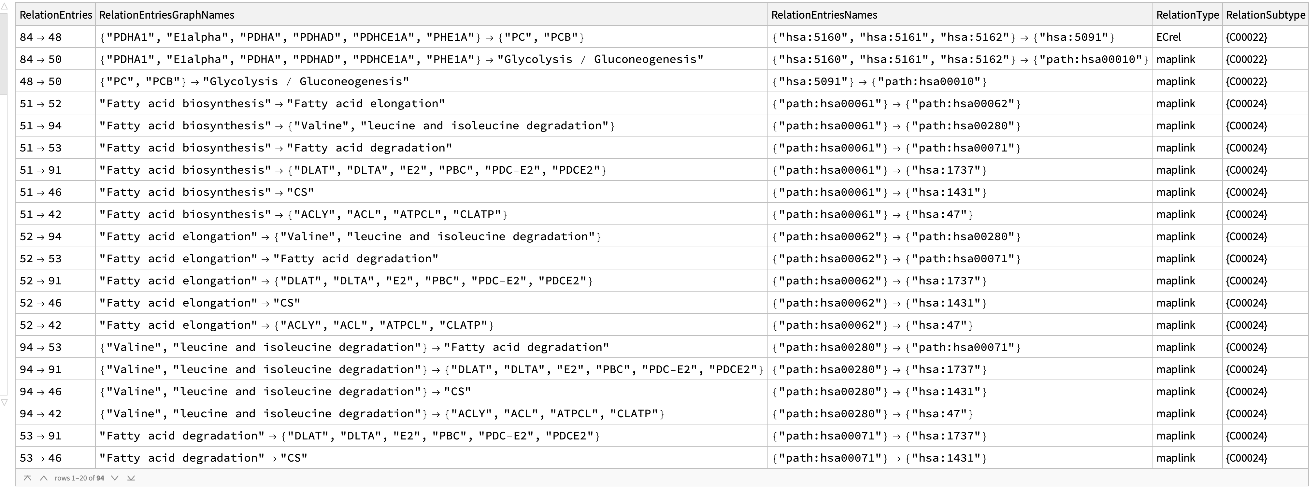 |
Get the reactions of the citrate cycle (TCA cycle) of a human:
| In[16]:= |
| Out[16]= | 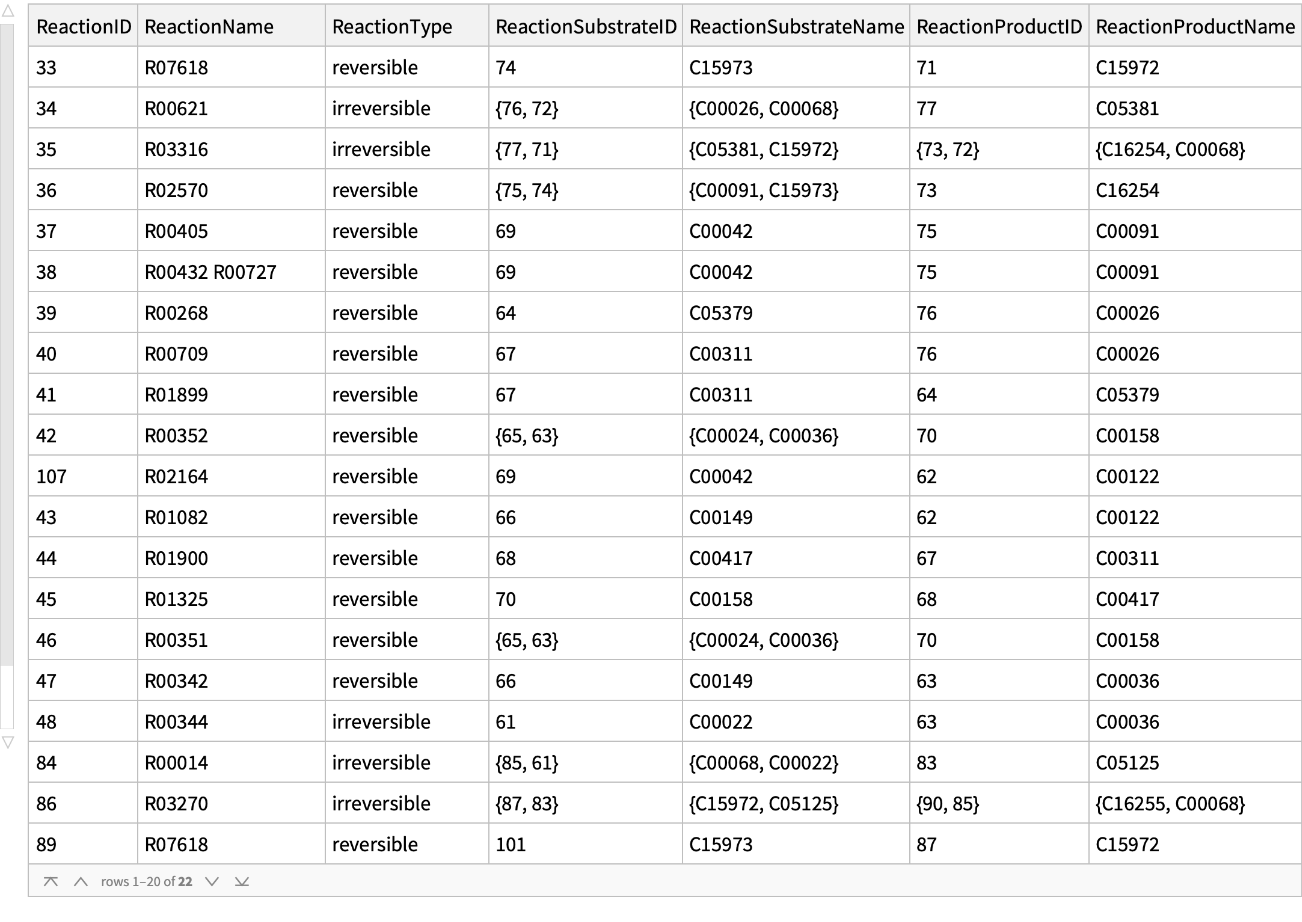 |
Get the Graph of the citrate cycle (TCA cycle) of a human. Get the complete names of the path components hovering over them. Click the vertex name and green boxes to get more information:
| In[17]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "HomoSapiens::4pydj"], "citrate cycle (TCA cycle)", "Graph", VertexSize -> Medium]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/489b9b3f490e1646.png) |
| Out[17]= | 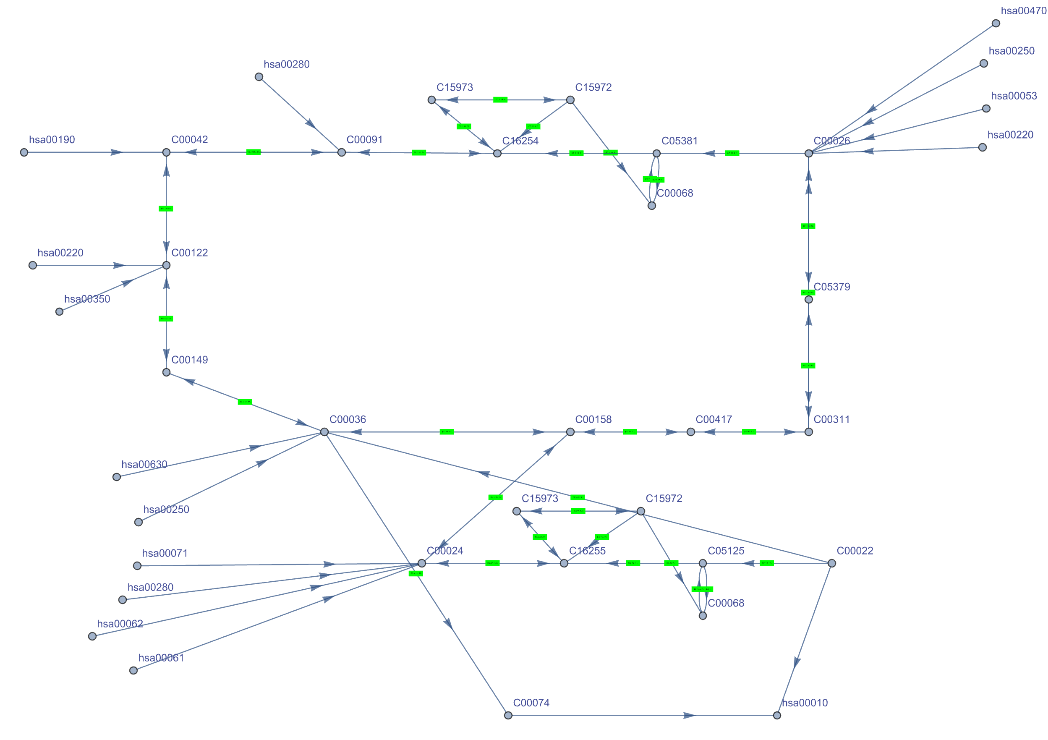 |
Get the Graph of the p53 signaling pathway of a macaque. Get the complete names of the path components hovering over them. Click the vertex name to get more information:
| In[18]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "MacacaFascicularis::3cnwm"], "p53 signaling pathway", "Graph", VertexSize -> {0.015, .008}, VertexStyle -> Hue[0.31, 0.35, 1], VertexLabelStyle -> Directive[FontFamily -> "Arial", 8]]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/7b33a429e7ddac63.png) |
| Out[18]= | 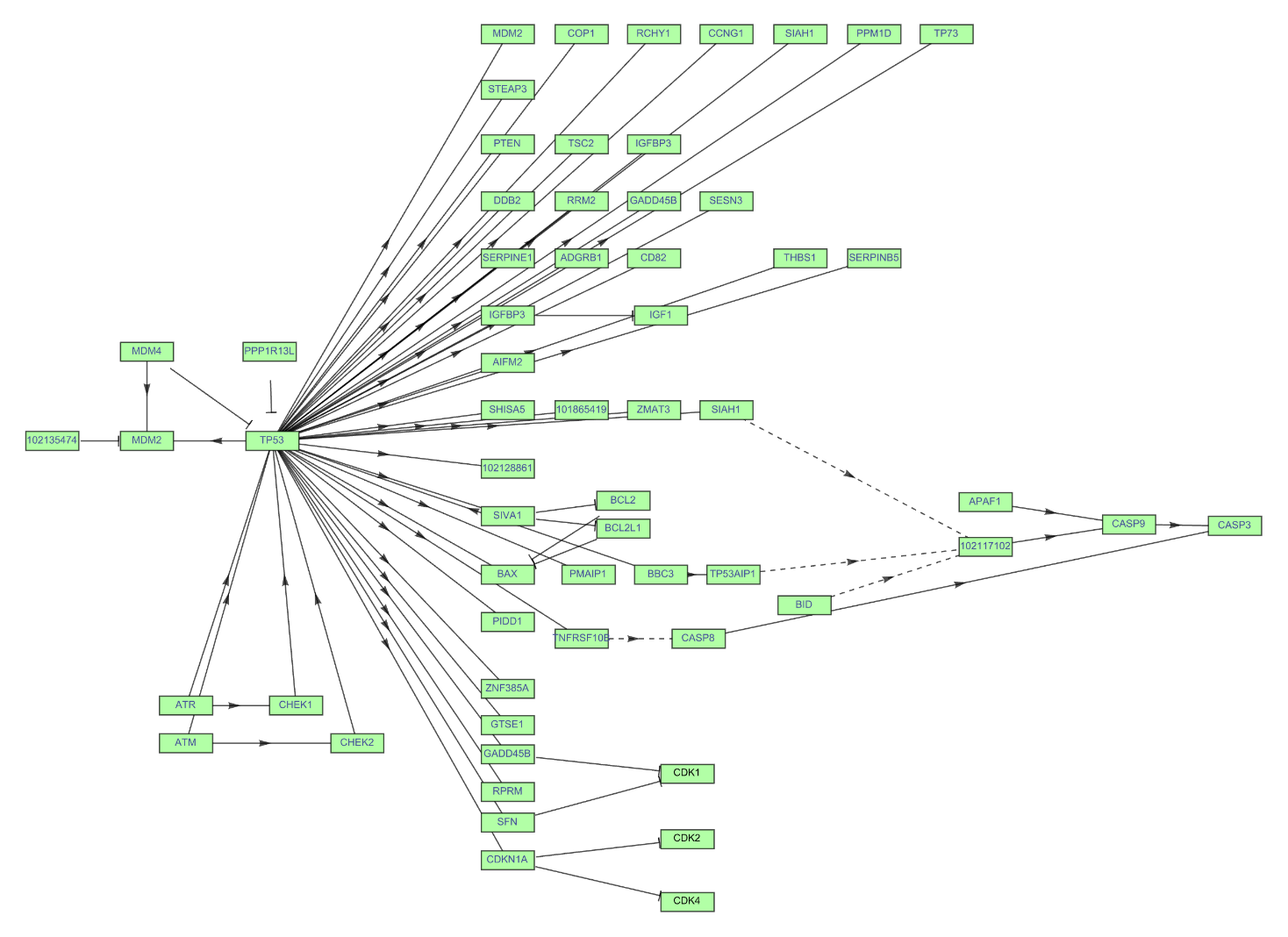 |
Get the Graph of the carbon metabolism of a human. Get the complete names of the graph components hovering over them. Click the vertex name and edges to get more information:
| In[19]:= |
| Out[19]= | 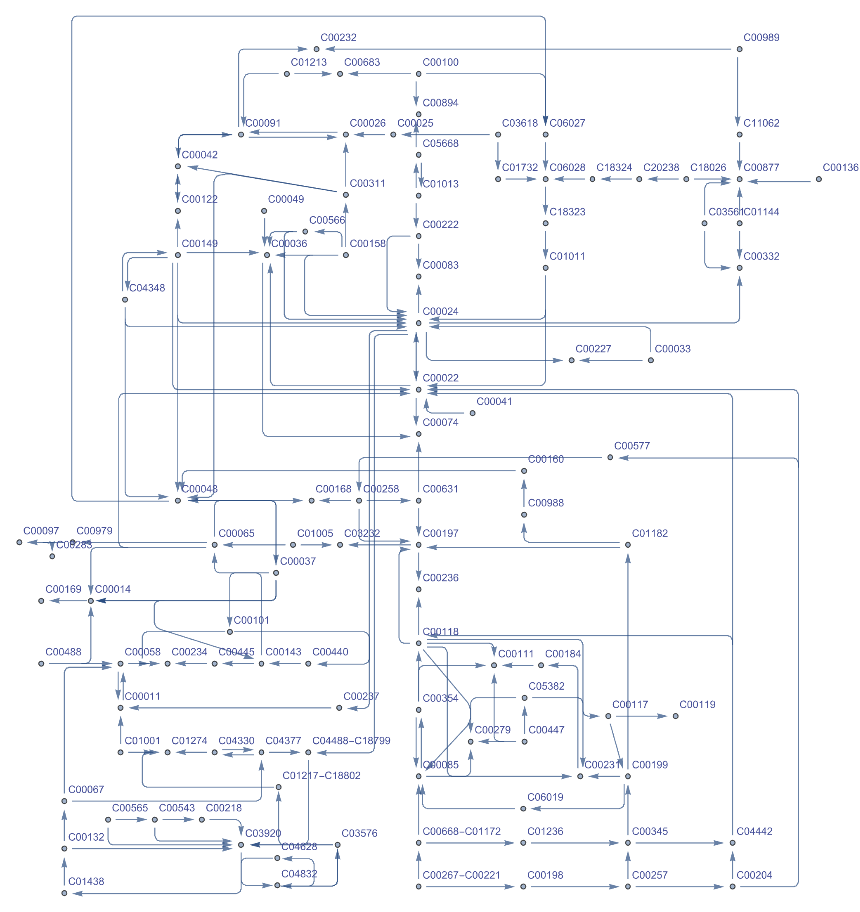 |
Get information of BID in pathway "04115" of "mcf":
| In[20]:= |
| Out[20]= |  |
Get information of all the relations in which CASP9 made part of in pathway "04115" of "mcf":
| In[21]:= |
Get information of the relation CASP9 -> CASP3 in pathway "04115" of "mcf":
| In[22]:= |
| Out[22]= |  |
Get information of all reactions in which C00631 is a part of the pathway "01200" of "hsa":
| In[23]:= |
Get information of the reaction CASP9 -> CASP3 in pathway "04115" of "mcf":
| In[24]:= |
| Out[24]= |  |
You may change VertexSize, but it will affect the view of the edges:
| In[25]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "MacacaFascicularis::3cnwm"], "p53 signaling pathway", "Graph", VertexSize -> {0.02, .008}, VertexStyle -> Hue[0.31, 0.35, 1], VertexLabelStyle -> Directive[FontFamily -> "Arial", 8]]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/4ab59515ffce2cc8.png) |
| Out[25]= | 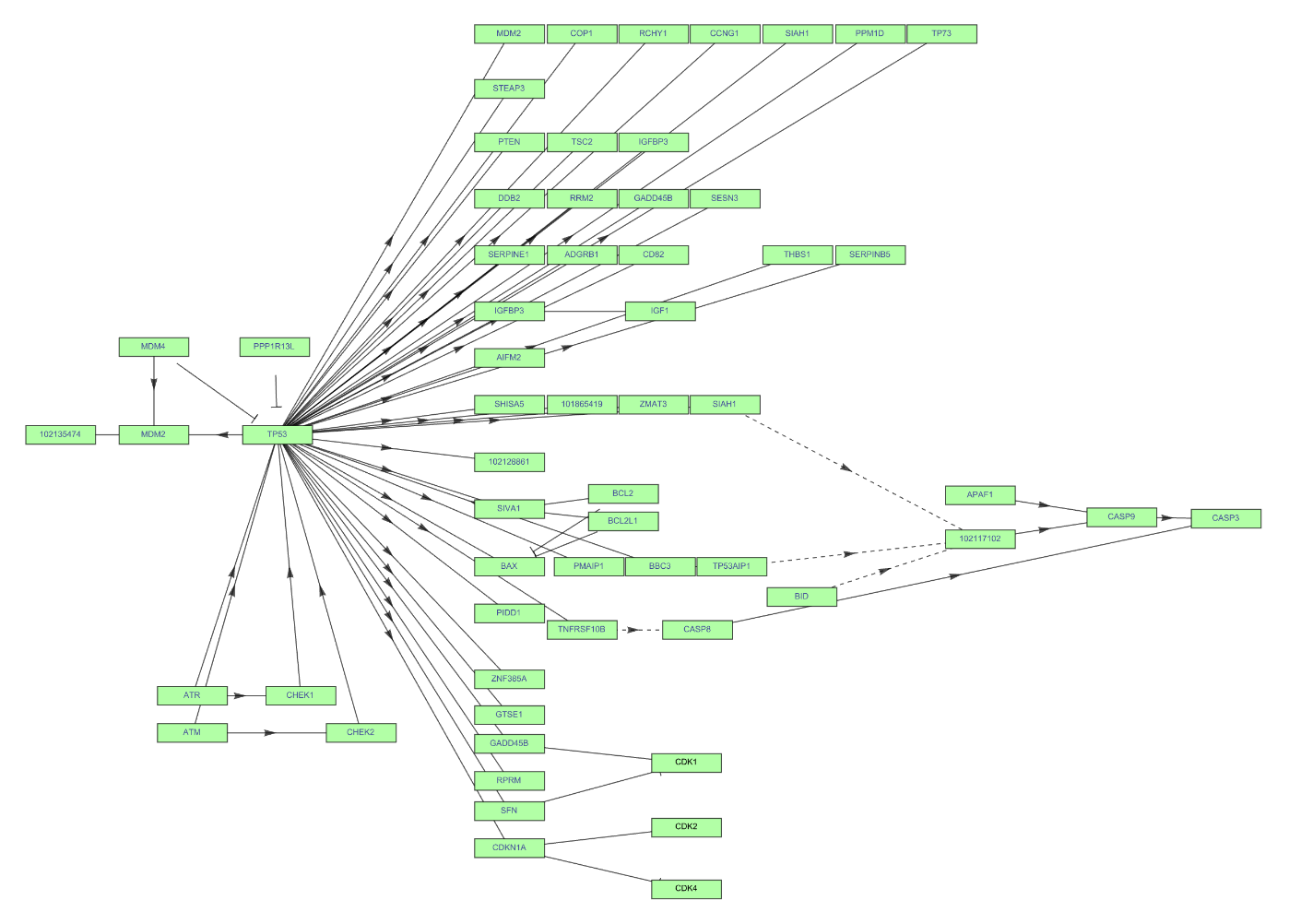 |
Make adjustments to better display the graph:
| In[26]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "MacacaFascicularis::3cnwm"], "p53 signaling pathway", "Graph", VertexSize -> {0.02, .008}, VertexStyle -> Hue[0.31, 0.35, 1], VertexLabelStyle -> Directive[FontFamily -> "Arial", 8], "Distance" -> 0.025]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/654ae55f72ec3c14.png) |
| Out[26]= | 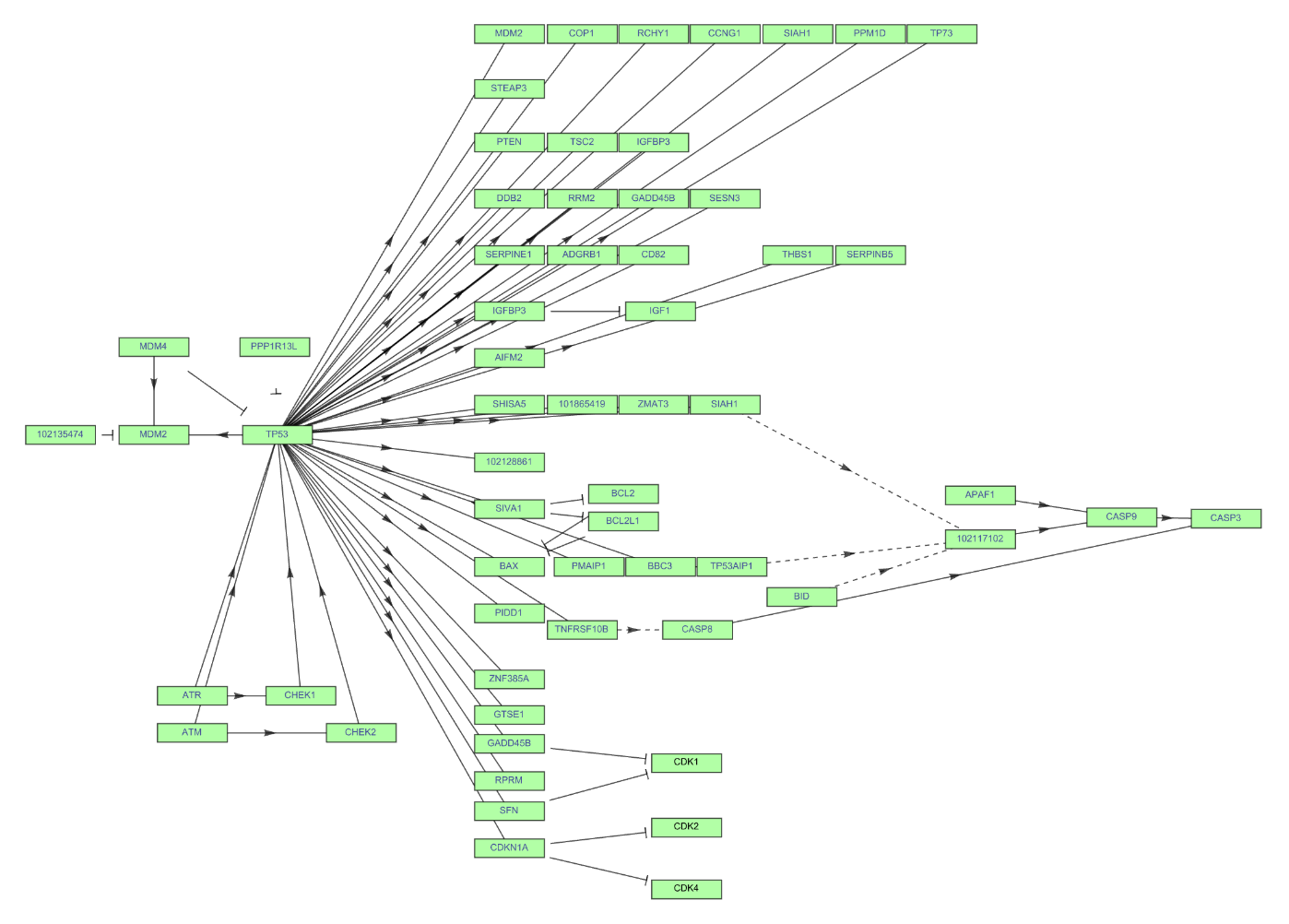 |
Highlight vertices base on their EntryID or any EntryGraphName in the graph of a KEGG pathway:
| In[27]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "MacacaFascicularis::3cnwm"], "p53 signaling pathway", "Graph", VertexSize -> {0.015, .008}, VertexStyle -> Hue[0.31, 0.35, 1], VertexLabelStyle -> Directive[FontFamily -> "Arial", 8], GraphHighlight -> {"CCND3", 10}]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/66d1d3f9264b9ce0.png) |
| Out[27]= | 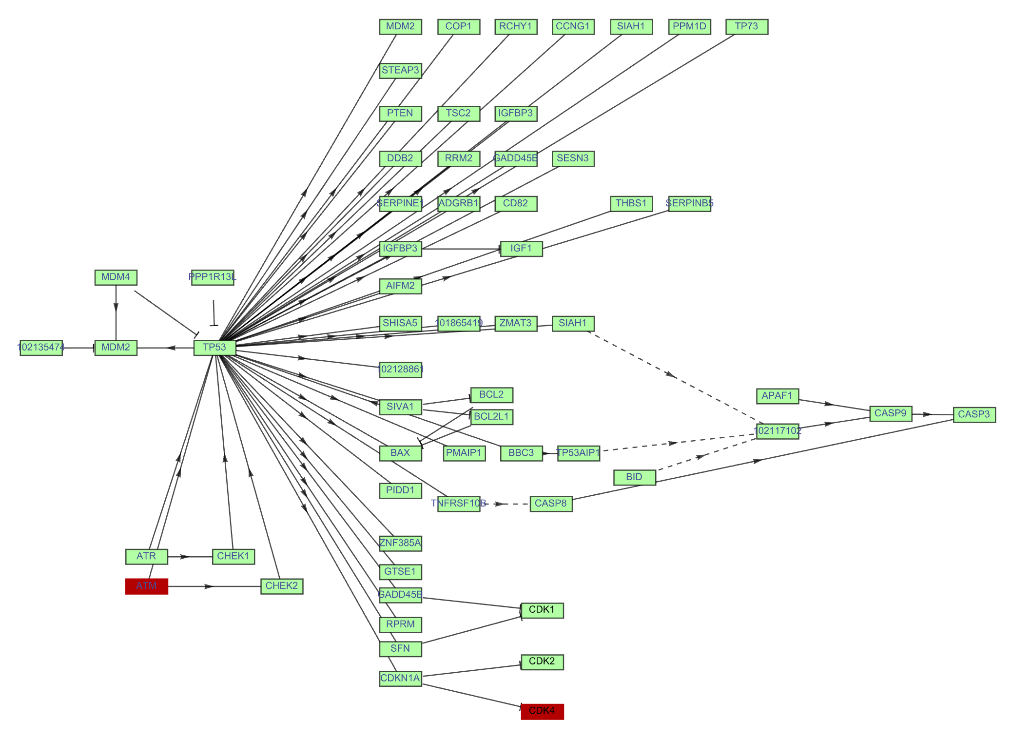 |
Highlight some edges and vertices in the graph of a KEGG pathway:
| In[28]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "MacacaFascicularis::3cnwm"], "p53 signaling pathway", "Graph", VertexSize -> {0.015, .008}, VertexStyle -> Hue[0.31, 0.35, 1], VertexLabelStyle -> Directive[FontFamily -> "Arial", 8], GraphHighlight -> {10 \[DirectedEdge] 9, 10, 9, "CASP8" \[DirectedEdge] "CASP3"}]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/272d262da78b0e2b.png) |
| Out[28]= | 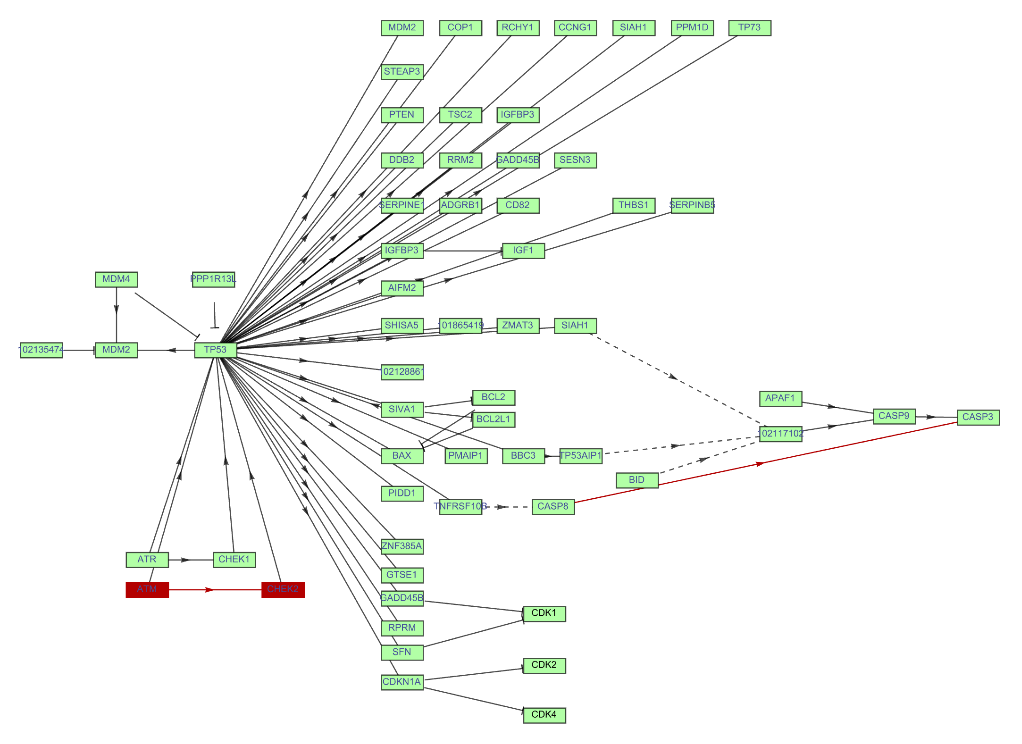 |
Change the style to highlight edges and vertices in the graph of a KEGG pathway:
| In[29]:= | ![ResourceFunction["KEGGPathway"][
Entity["TaxonomicSpecies", "MacacaFascicularis::3cnwm"], "p53 signaling pathway", "Graph", VertexSize -> {0.015, .008}, VertexLabelStyle -> Directive[FontFamily -> "Arial", 8], VertexStyle -> {"CASP8" -> Yellow, "CASP3" -> Yellow, Hue[0.31, 0.35, 1]}, EdgeStyle -> {"CASP8" \[DirectedEdge] "CASP3" -> Gray}]](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/779ddb9a192ea2ce.png) |
| Out[29]= | 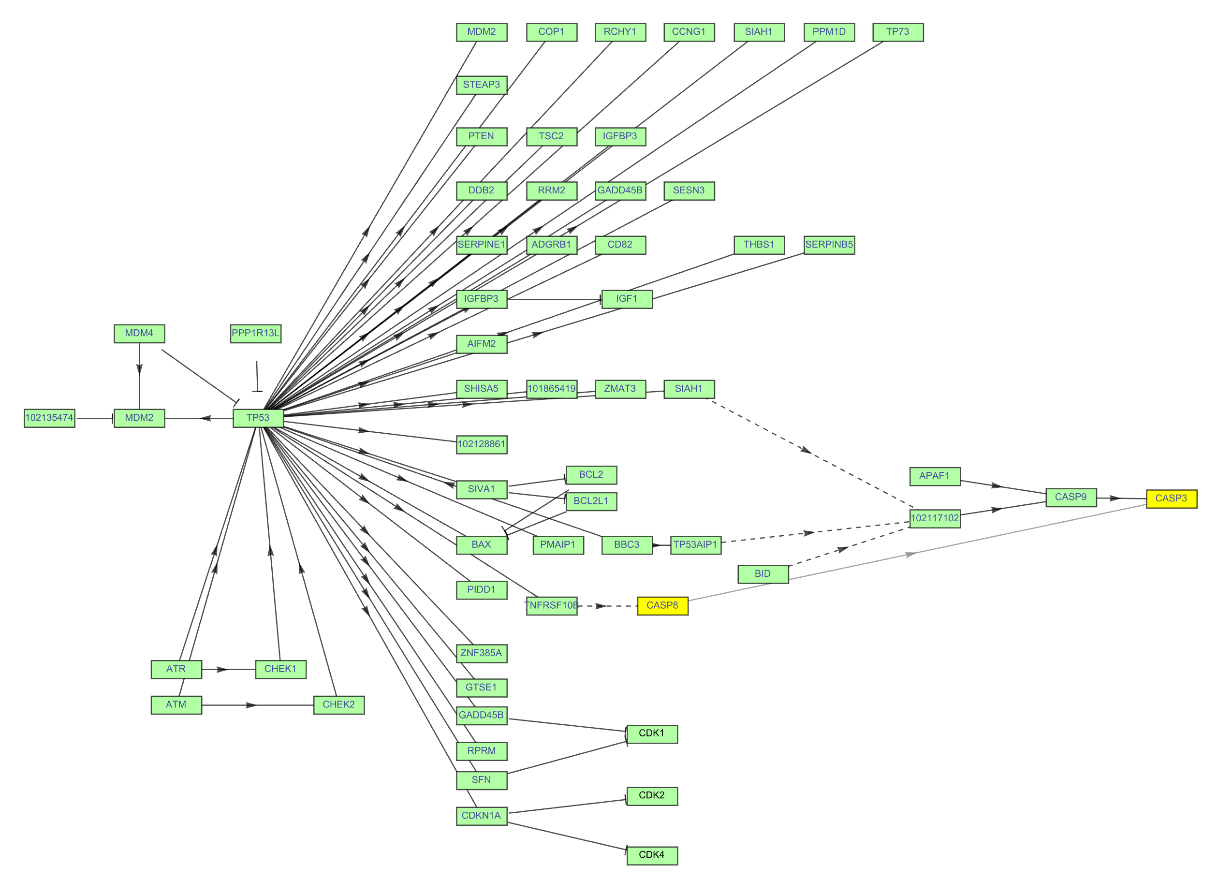 |
Get the vertex list of "mcf" / "04115" entries:
| In[30]:= | ![vNames = Normal[ResourceFunction["KEGGPathway"]["mcf", "04115", "Entries"][
All, #EntryID -> #EntryGraphName &]];
graph = ResourceFunction["KEGGPathway"]["mcf", "04115", "Graph"];](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/1a256f6bedaf8215.png) |
| In[31]:= |
| Out[31]= |  |
Get the edge list:
| In[32]:= |
| Out[32]= |  |
Get the number of vertices:
| In[33]:= |
| Out[33]= |
Get the number of edges:
| In[34]:= |
| Out[34]= |
Get the vertex degrees:
| In[35]:= |
| Out[35]= |  |
Make a histogram:
| In[36]:= |
| Out[36]= |  |
Get the vertex out-degree list:
| In[37]:= |
| Out[37]= |  |
Make a histogram:
| In[38]:= |
| Out[38]= |  |
Get the vertex in-degree list:
| In[39]:= |
| Out[39]= |  |
Make a histogram:
| In[40]:= |
| Out[40]= |  |
Get the BetweennessCentrality:
| In[41]:= |
| Out[41]= |  |
Make a histogram:
| In[42]:= |
| Out[42]= | 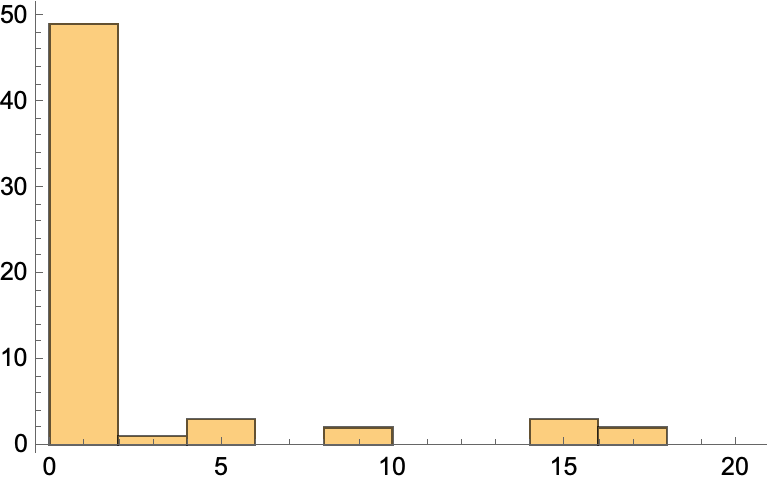 |
KEGGPathway fails if given the wrong organism name or path code:
| In[43]:= |
| Out[43]= |
KEGGPathway fails if given an Entity that does not match a organism name:
| In[44]:= |
| Out[44]= |
Get a cluster analysis of the graphs based on some network properties:
| In[45]:= | ![org = "hsa";
pathCode = {"03440", "03460", "04010", "04012", "04929", "04912", "04917"};
titles = ResourceFunction["KEGGPathway"][org, #, "Information"][[;; 2]] & /@ pathCode;
gs = ResourceFunction["KEGGPathway"][org, #, "GraphRelation"] & /@ pathCode;
gs2 = MapThread[
Tooltip[Graph[EdgeList[#] 1, ImageSize -> Tiny, Frame -> True], #2] &, {gs, titles}];
c = {AcyclicGraphQ[#], BipartiteGraphQ[#], PlanarGraphQ[#], VertexCount[#], EdgeCount[#], GraphDensity[#], GlobalClusteringCoefficient[#], GraphAssortativity[#]} & /@ gs;](https://www.wolframcloud.com/obj/resourcesystem/images/c26/c26fb78f-9391-4e5b-8348-1b3a9bcb8293/1-0-0/676a9557b53272f1.png) |
See a dendrogram of the resulting graphs:
| In[46]:= |
| Out[47]= | 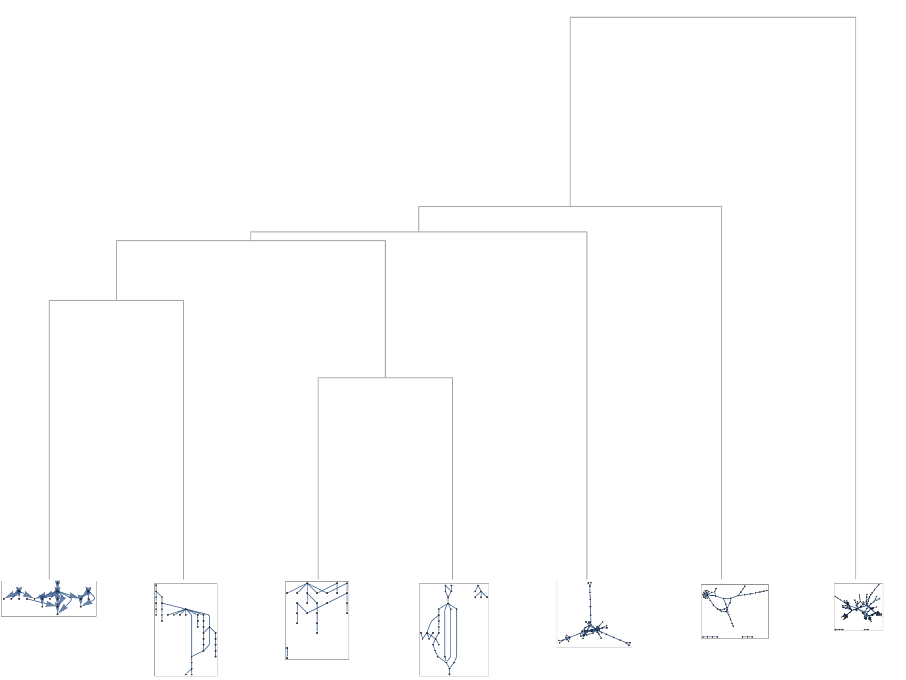 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License