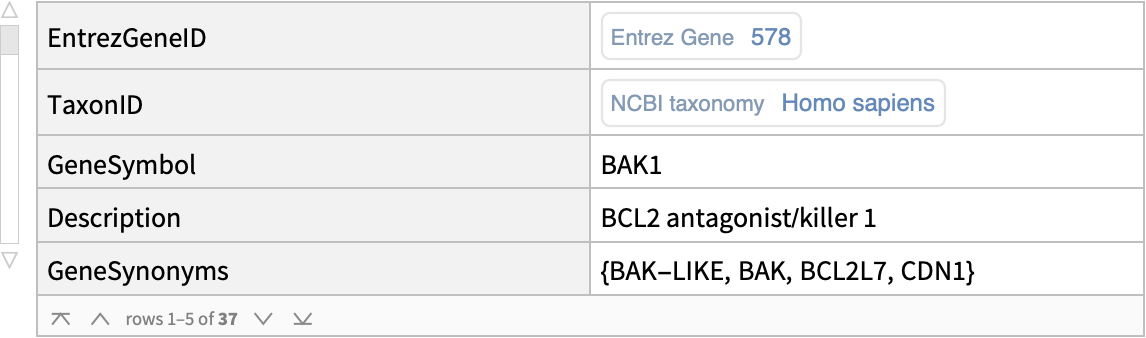
| "EntrezGeneID" | identifier for a gene as assigned by the Entrez Gene project |
| "TaxonID" | identifier for the NCBI Taxonomy database |
| "GeneSymbol" | symbolic representation of a gene |
| "Description" | detailed description of a gene |
| "GeneSynonyms" | gene symbol aliases |
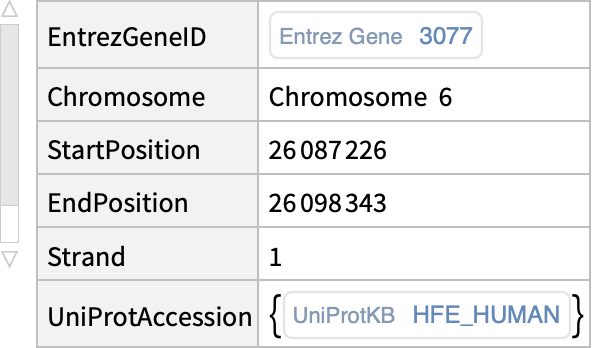
| "Chromosome" | chromosome of a gene |
| "ChromosomeBand" | cytoband location of a gene |
| "StartPosition" | starting sequence position of a gene |
| "EndPosition" | ending sequence position of a gene |
| "Strand" | coding strand of a gene |
| "NCBIHomologGeneIDs" | identifier for associated NCBI Taxonomy and Entrez Gene |
| "EnsemblGeneID" | identifier for a gene as assigned by Ensembl project |
| "EnsemblBiotype" | biotype of a gene or a protein as assigned by Ensembl project |
| "EnsemblTranscriptID" | identifier for a transcript as assigned by Ensembl project |
| "EnsemblProteinID" | identifier for a protein as assigned by Ensembl project |
| "EnsemblHomologGeneIDs" | identifier for associated NCBI Taxonomy and Ensembl genes |
| "EnsemblHomologProteinIDs" | identifier for associated NCBI Taxonomy and Ensembl proteins |
| "RefSeqGenomicAccession" | genomic accession for RefSeq |
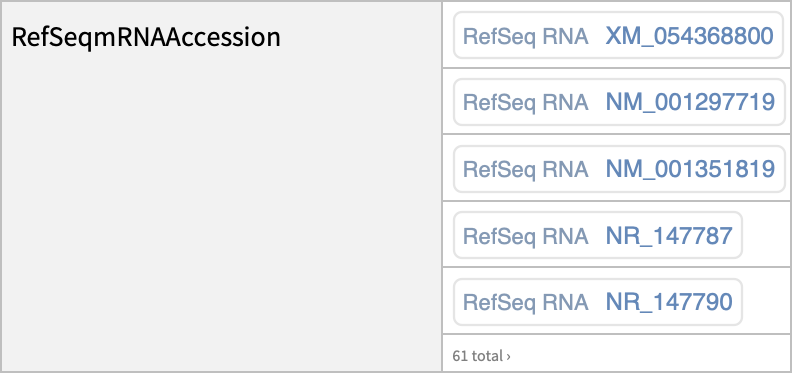
| "RefSeqmRNAAccession" | mRNA accession for RefSeq |
| "RefSeqncRNAAccession" | non-coding RNA accession for RefSeq |
| "RefSeqProteinAccession" | protein accession for RefSeq |
| "GenBankNucleotideAccession" | mRNA accession for the GenBank database |
| "GenBankProteinAccession" | protein accession for the GenBank database |
| "PDBID" | identifier for RCSB PDB (Research Collaboratory for Structural Bioinformatics Protein Data Bank) |
| "UniProtAccession" | identifier for UniProt (Universal Protein Resource) sequences |
| "GOBiologicalProcess" | identifier for Gene Ontology (GO) associated with biological processes |
| "GOCellularComponent" | identifier for Gene Ontology (GO) associated with cellular components |
| "GOMolecularFunction" | identifier for Gene Ontology (GO) associated with molecular functions |
| "CPDBProteinInteractor" | UniProt entry name of the interacting protein from ConsensusPathDB |
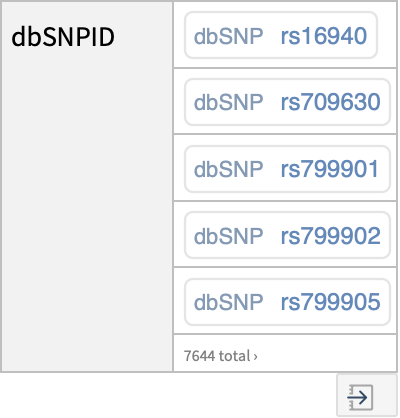
| "dbSNPID" | identifier for the dbSNP (Single Nucleotide Polymorphism database) |
| "CTDDiseaseInfo" | associated disease based on CTD (Comparative Toxicogenomics Database) |
| "DrugBankDrugID" | identifier for associated drugs from DrugBank database |
| "KEGGDiseaseID" | identifier for the associated disease in KEGG (Kyoto Encyclopedia of Genes and Genomes) database |
| "KEGGGeneID" | identifier for KEGG gene |
| "KEGGPathwayID" | identifier for the associated KEGG pathway |
| "ReactomeID" | identifier for the associated Reactome pathway |
| "PubMedID" | identifier for the associated PubMed articles |
![ResourceFunction[
"BioDBnetGeneData"]["HFE1", {"EntrezGeneID", "Chromosome", "StartPosition", "EndPosition", "Strand", "UniProtAccession"}]](https://www.wolframcloud.com/obj/resourcesystem/images/73f/73f25ec5-bf6c-435a-8087-74a79d984422/2c2a7bfd3bddba71.png)
![ResourceFunction[
"BioDBnetGeneData"][{"swiss cheese", "timeless"}, {"GeneSymbol", "EnsemblGeneID", "GOBiologicalProcess", "KEGGPathwayID"}, {"Species" -> Entity["TaxonomicSpecies", "DrosophilaMelanogaster::ynxq4"]}]](https://www.wolframcloud.com/obj/resourcesystem/images/73f/73f25ec5-bf6c-435a-8087-74a79d984422/2d9fbe71b479004a.png)



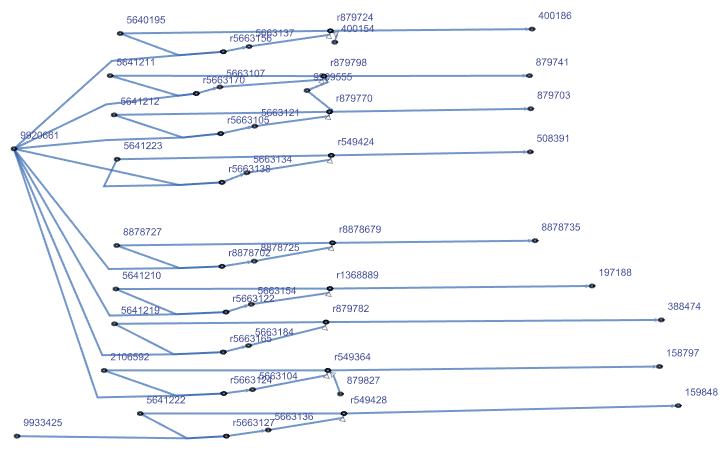
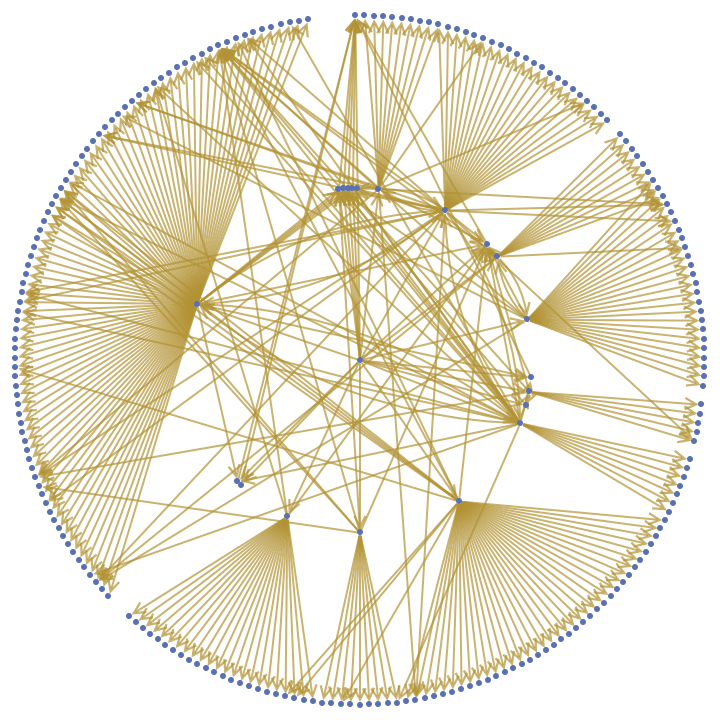
![NestGraph[(Flatten[
Normal@Values[
ResourceFunction["BioDBnetGeneData"][StringTrim[#, "_" ~~ __], "CPDBProteinInteractor"]], 1] /. {l_List} :> l) &, "BAK1", 2, VertexLabels -> Placed["Name", Tooltip], GraphStyle -> "LargeNetwork",
GraphLayout -> "RadialEmbedding"]](https://www.wolframcloud.com/obj/resourcesystem/images/73f/73f25ec5-bf6c-435a-8087-74a79d984422/4dc4315030a2bc53.png)