Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Calculate the free energy changes for folding one RNA strand using nearest neighbor parameters
ResourceFunction["RNAFoldingFreeEnergyChange"][biosequence] calculates the free energy change for one folded RNA BioSequence strand. | |
ResourceFunction["RNAFoldingFreeEnergyChange"][biosequence, options] calculates the free energy change for one folded RNA BioSequence strand with optional insights. |
| Automatic | calculate free energy change |
| "Structures" | identify substructures with connected indices |
| "StructuresWithEnergy" | identify substructures with connected indices, and calculate their respective free energy changes |
Predict the free energy change for a given folded BioSequence:
| In[1]:= |
| Out[1]= |
Visualize the BioSequence with BioSequencePlot:
| In[2]:= |
| Out[2]= | 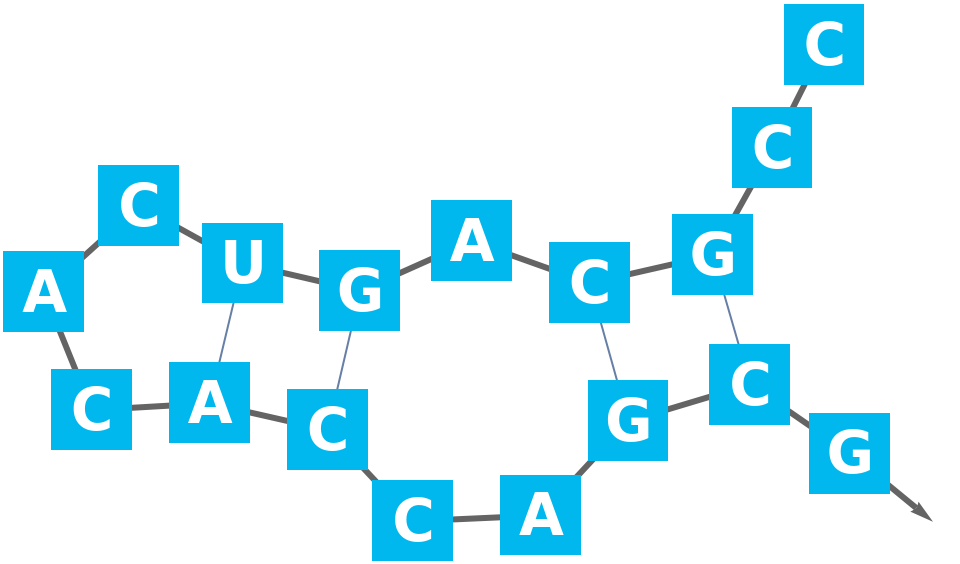 |
Return a breakdown of the various structures:
| In[3]:= | ![ResourceFunction["RNAFoldingFreeEnergyChange"][
BioSequence["RNA", "CCGCAGUCACACCAGCG", Bond /@ {{3, 16}, {4, 15}, {6, 12}, {7, 11}}], Method -> "Structures"]](https://www.wolframcloud.com/obj/resourcesystem/images/709/709cb6ed-994e-48ff-8d2b-430f8a45eb24/4ed336b4cf9aa899.png) |
| Out[3]= |
Predict respective free energy changes for the various structures:
| In[4]:= | ![ResourceFunction["RNAFoldingFreeEnergyChange"][
BioSequence["RNA", "CCGCAGUCACACCAGCG", Bond /@ {{3, 16}, {4, 15}, {6, 12}, {7, 11}}], Method -> "StructuresWithEnergy"]](https://www.wolframcloud.com/obj/resourcesystem/images/709/709cb6ed-994e-48ff-8d2b-430f8a45eb24/28dc2b6a3e334d3e.png) |
| Out[4]= | 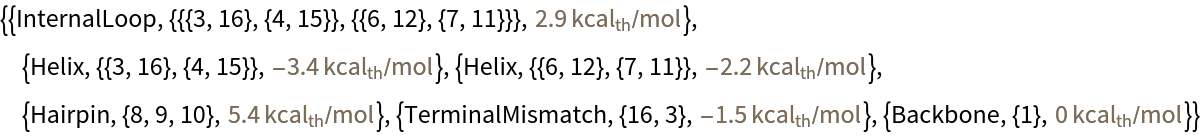 |
| In[5]:= |
| Out[5]= |
Possible base pairs can yield invalid nearest neighbor structures and ultimately in an Indeterminate result:
| In[6]:= |
| Out[6]= |
A more detailed look:
| In[7]:= |
| Out[7]= |
Visual inspection of the BioSequence with Graph:
| In[8]:= | ![With[{len = StringLength[#[[2]]]}, Graph[Labeled[#, #] & /@ Range[len], DeleteDuplicates[
Join[(# <-> (# + 1)) & /@ Range[len - 1], (#1 <-> #2) & @@@ #[[3, All, 1]]]], GraphLayout -> {"SpringEmbedding", "Multilevel" -> "MaximalIndependentVertexSet"}]] &@
BioSequence["RNA", "CCGCAGUCACACCAGCG", Bond /@ {{4, 6}}]](https://www.wolframcloud.com/obj/resourcesystem/images/709/709cb6ed-994e-48ff-8d2b-430f8a45eb24/4936b108cdebf4c0.png) |
| Out[8]= |  |
Pseudoknots are not supported:
| In[9]:= | ![ResourceFunction["RNAFoldingFreeEnergyChange"][
BioSequence["RNA", "CUCGUCUAGUCAUUUCUGGCCCCACUGGAGGUCGAG", Bond /@ {{4, 20}, {5, 19}, {8, 17}, {9, 16}, {14, 35}, {15, 34}, {22, 28}, {23, 27}}]]](https://www.wolframcloud.com/obj/resourcesystem/images/709/709cb6ed-994e-48ff-8d2b-430f8a45eb24/0e9a9fb5f9b268ac.png) |
| Out[9]= |
Closer inspection of the respective BioSequence:
| In[10]:= | ![With[{len = StringLength[#[[2]]]}, Graph[Labeled[#, #] & /@ Range[len], DeleteDuplicates[
Join[(# <-> (# + 1)) & /@ Range[len - 1], (#1 <-> #2) & @@@ #[[3, All, 1]]]], GraphLayout -> {"SpringEmbedding", "Multilevel" -> "MaximalIndependentVertexSet"}]] &@
BioSequence["RNA", "CUCGUCUAGUCAUUUCUGGCCCCACUGGAGGUCGAG", Bond /@ {{4, 20}, {5, 19}, {8, 17}, {9, 16}, {14, 35}, {15, 34}, {22, 28}, {23, 27}}]](https://www.wolframcloud.com/obj/resourcesystem/images/709/709cb6ed-994e-48ff-8d2b-430f8a45eb24/24b469eb3558c77d.png) |
| Out[10]= | 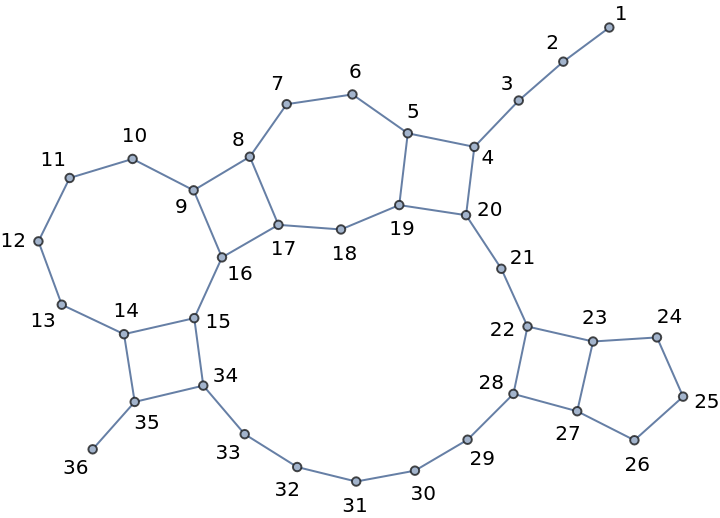 |
This work is licensed under a Creative Commons Attribution 4.0 International License