Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Build a matrix containing which biomolecular sequence positions can bond
ResourceFunction["BioSequenceFoldingMatrix"][bioseq] obtains a matrix of all of the positions which a biological sequence bioseq is allowed to form a bond. |
| "AdditionalBondingPairs" | {} | specify additional complementary pairs |
| "UseTypeRules" | True | determine if "DNA" or "RNA" terms will bond to their complement letters |
Build the folding matrix based on built-in DNA complementation rules:
| In[1]:= |
| Out[1]= | 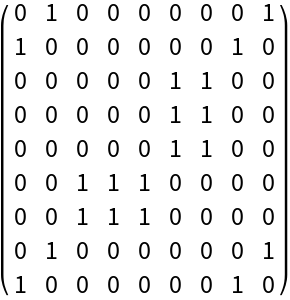 |
Build a folding matrix for RNA that includes additional "wobble" pairings between "G" and "U":
| In[2]:= |
| Out[2]= | 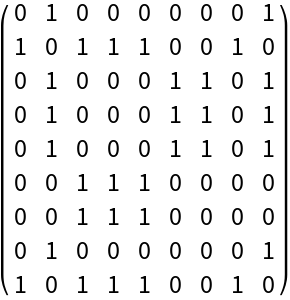 |
See only the wobble pairings by turning off the rules from the type:
| In[3]:= |
| Out[3]= | 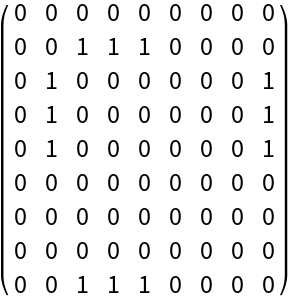 |
Find all potential bonding pairs:
| In[4]:= | ![Cases[ArrayRules[
ResourceFunction["BioSequenceFoldingMatrix"][
BioSequence["RNA", "CGUUUAACG", {}], "AdditionalBondingPairs" -> {{"G", "U"}}]], HoldPattern[Rule[x_, 1]] :> x]](https://www.wolframcloud.com/obj/resourcesystem/images/514/5145ef7c-5f14-4726-a598-646fbc35be27/4d2270a7cfcbb433.png) |
| Out[4]= |
This work is licensed under a Creative Commons Attribution 4.0 International License