Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Import protein data in the Protein Data Bank (PDB) format
ResourceFunction["PDBImport"][src] applies a PDB import to the source src. | |
ResourceFunction["PDBImport"][src,"String"] imports protein data from the source src as a string of PDB format. | |
ResourceFunction["PDBImport"][src, "AtomAssociation"] imports a protein atom Association from the source src in the RCSB Protein Data Bank format. | |
ResourceFunction["PDBImport"][src,row] imports a protein atom starting from the specified row. | |
ResourceFunction["PDBImport"][provider→identifier,…] imports the protein data from the given provider with the identifer appropriate to that provider. |
Import a PDB file corresponding to an RCSB ID:
| In[1]:= |
| Out[1]= | 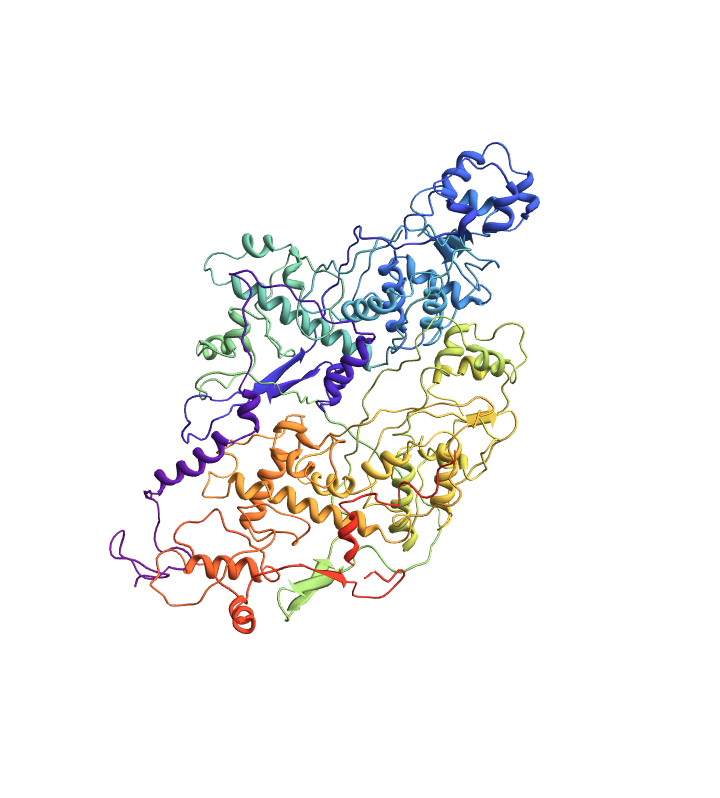 |
Import an Association of atomic properties corresponding to an RCSB ID:
| In[2]:= |
| Out[2]= |  |
Extract the x-coordinates:
| In[3]:= |
| Out[3]= |
Extract the atom types:
| In[4]:= |
| Out[4]= |
Extract the atomic masses:
| In[5]:= |
| Out[5]= |
Import an RCSB data file starting from row 154:
| In[6]:= |
| Out[6]= | 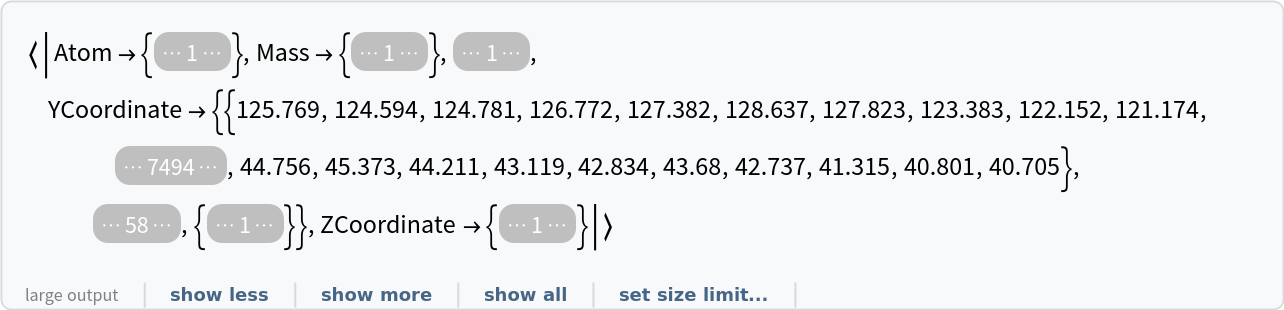 |
Extract the x-coordinates:
| In[7]:= |
| Out[7]= |
Extract the atom types:
| In[8]:= |
| Out[8]= |
Import an RCSB data file as a PDB string:
| In[9]:= |
| Out[9]= |  |
Extract the x-coordinates using PDB import (which converts to picometers):
| In[10]:= |
| Out[10]= |
Extract the atom types:
| In[11]:= |
| Out[11]= |
Extract a protein chain and calculate the center of mass of its subunits:
| In[12]:= |
| In[13]:= | ![centermass[mass_, coor_] := Sum[mass[[i]]* coor[[i]], {i, Length[mass]}] /
Sum[mass[[i]], { i, Length[mass]}]](https://www.wolframcloud.com/obj/resourcesystem/images/e30/e304aadb-6c4b-41a1-a7d2-8c1e941054bc/7004147e5911d337.png) |
| In[14]:= | ![xset = MapThread[centermass, {data["Mass"], data["XCoordinate"]}];
yset = MapThread[centermass, {data["Mass"], data["YCoordinate"]}];
zset = MapThread[centermass, {data["Mass"], data["ZCoordinate"]}];](https://www.wolframcloud.com/obj/resourcesystem/images/e30/e304aadb-6c4b-41a1-a7d2-8c1e941054bc/0a3f27456d89d45a.png) |
| In[15]:= |
Plot the center of mass of the subunits:
| In[16]:= |
| Out[16]= | 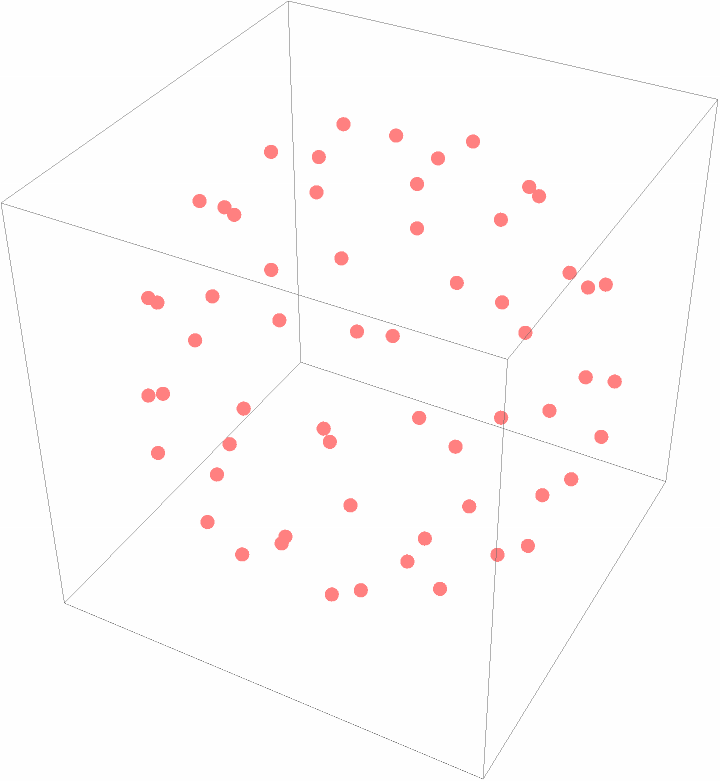 |
This work is licensed under a Creative Commons Attribution 4.0 International License