Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get pathways and additional information from the Reactome database
ResourceFunction["ReactomePathways"]["Organisms"] gives the dataset with basic information for all the organisms in Reactome. | |
ResourceFunction["ReactomePathways"]["ExternalDatabases"] gives the dataset with all the external databases of Reactome. | |
ResourceFunction["ReactomePathways"][spcode,"PrimaryPathways"] gives the top level pathways for a specific spcode in Reactome. | |
ResourceFunction["ReactomePathways"][pathcode,prop] gives a property prop for a specific pathcode in Reactome. | |
ResourceFunction["ReactomePathways"][keywords,"QueryPaths"] gives a dataset with pathways corresponding to keywords. | |
ResourceFunction["ReactomePathways"][externalidentifier,"QueryReactions"] gives a dataset with reactions corresponding to keywords. |
| "SecondaryPathways" | low level pathways inside a top level pathway |
| "ParentPathways" | all possible paths from the requested event to the top level pathway(s) |
| "Information" | general information of a pathway |
| "Compartments" | compartments of a pathway |
| "Entries" | biochemical components of a pathway |
| "Reactions" | reactions of a pathway |
| "Glyph" | information related to the nodes in the pathway |
| "Arc" | information related to the edges in the pathway |
| "Image" | get the Reactome Image of the pathway |
| "Graph" | get the graph of the pathway |
| "EntriesExternalIdentifiers" | list of participants in a given event and their external identifiers |
| "ArrowHeadSize" | 0.01 | set the arrowhead size |
| "Distance" | 8 | set the distance between the vertex and edge |
| "RemoveReactions" | False | delete vertices representing reactions |
| "Extension" | "png" | set the file extension to one of the following:png,jpg,jpeg,svg,or gif. |
| "Quality" | 10 | set the quality on a scale from 1 to 10 |
| "Flag" | None | gene name,protein or chemical identifier or Reactome identifier to highlight in the diagram |
| "FlagInteractors" | True | whether to take into account interactors for the flagging |
| "Title" | True | whether the name of the pathway is shown below |
| "Margin" | 0 | image margin from 0 to 20 |
| "Ehld" | True | whether textbook-like illustrations are taken into account (Electronic Help with Learning and Discovery) |
| "DiagramProfile" | "Modern" | Modern or Standard |
| "Token" | None | the analysis token with the results to be overlaid on top of the given diagram |
| "Resource" | "TOTAL" | the analysis resource for which the results will be overlaid on top of the given pathways overview |
| "AnalysisProfile" | "Standard" | Standard, Strosobar, or Copper Plus |
Get basic information for all the organisms in the database:
| In[1]:= |
| Out[2]= | 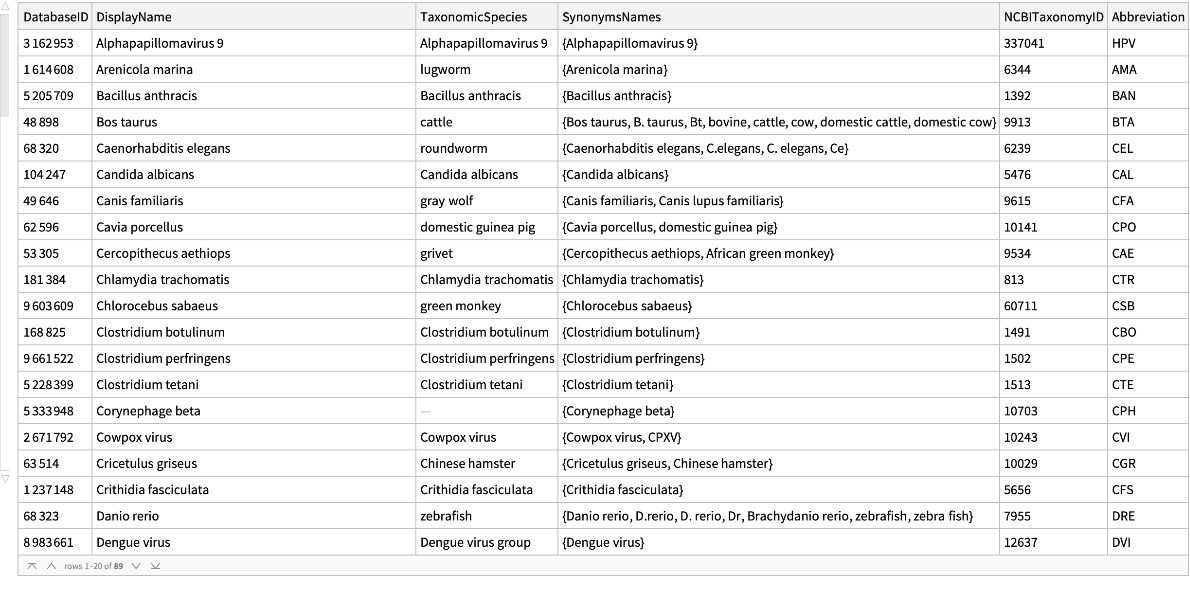 |
Databases of the external identifiers:
| In[3]:= |
| Out[162]= | 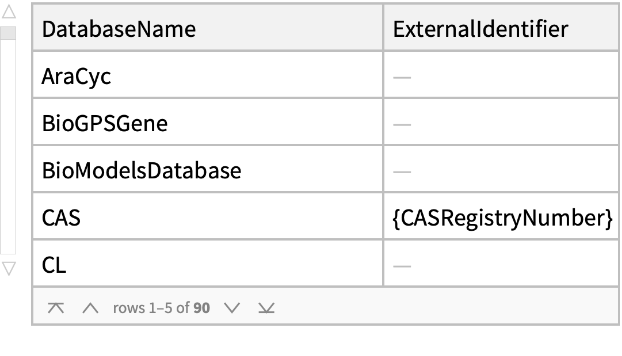 |
Get basic information for all the top pathways for Homo sapiens with NCBITaxonomyID 9606:
| In[163]:= |
| Out[164]= | 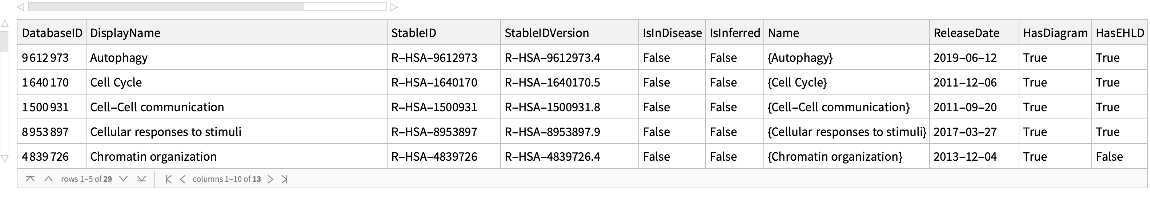 |
Get basic information for all the top pathways for Homo sapiens:
| In[165]:= |
| Out[165]= | 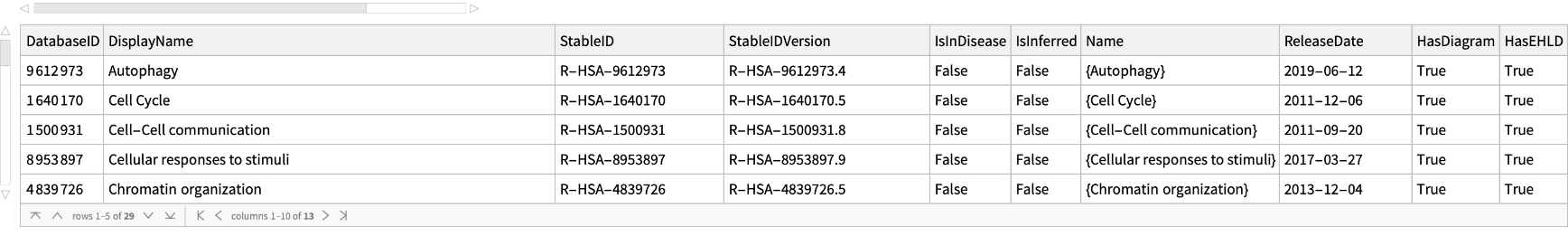 |
Get basic information for all the pathways in the primary pathway "R-HSA-180786" of Homo sapiens:
| In[166]:= |
| Out[166]= | 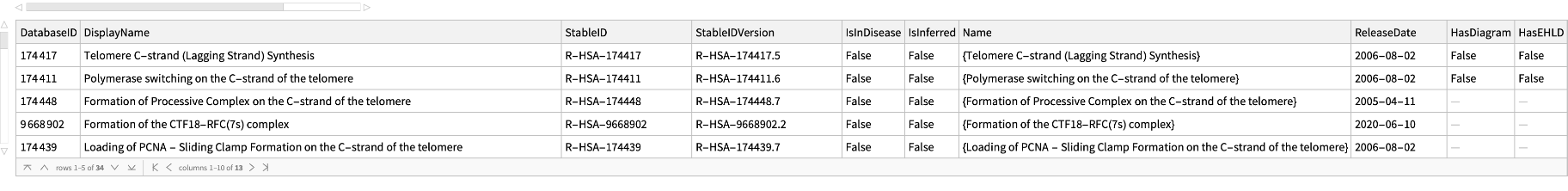 |
Get basic information for all the parent pathways of a pathway "R-HSA-5673001" of Homo sapiens:
| In[167]:= |
| Out[167]= | 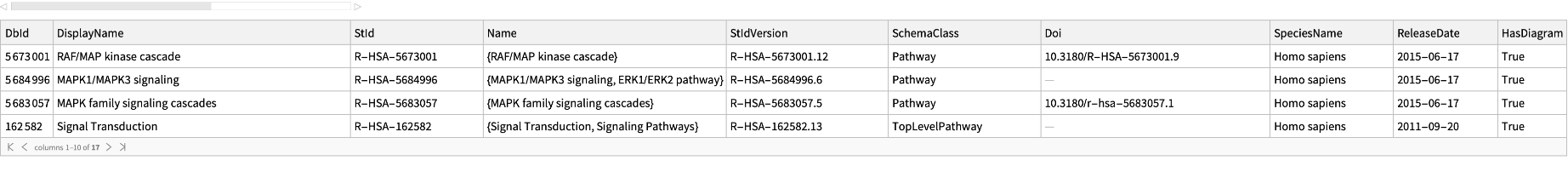 |
Get information of "R-HSA-180786" pathway for Homo sapiens:
| In[168]:= |
| Out[168]= |  |
Get the Entries of "R-HSA-180786" pathway for Homo sapiens:
| In[169]:= |
| Out[169]= | 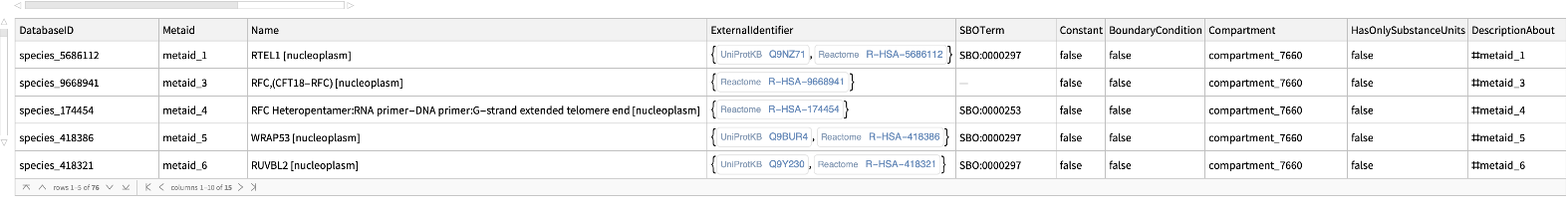 |
Get the Reactions of "R-HSA-180786" pathway for Homo sapiens:
| In[170]:= |
| Out[170]= |  |
Get the Graph of "R-HSA-180786" pathway for Homo sapiens. Get the complete names of the path components hovering over them. Click the vertex name to get more information:
| In[171]:= |
| Out[171]= | 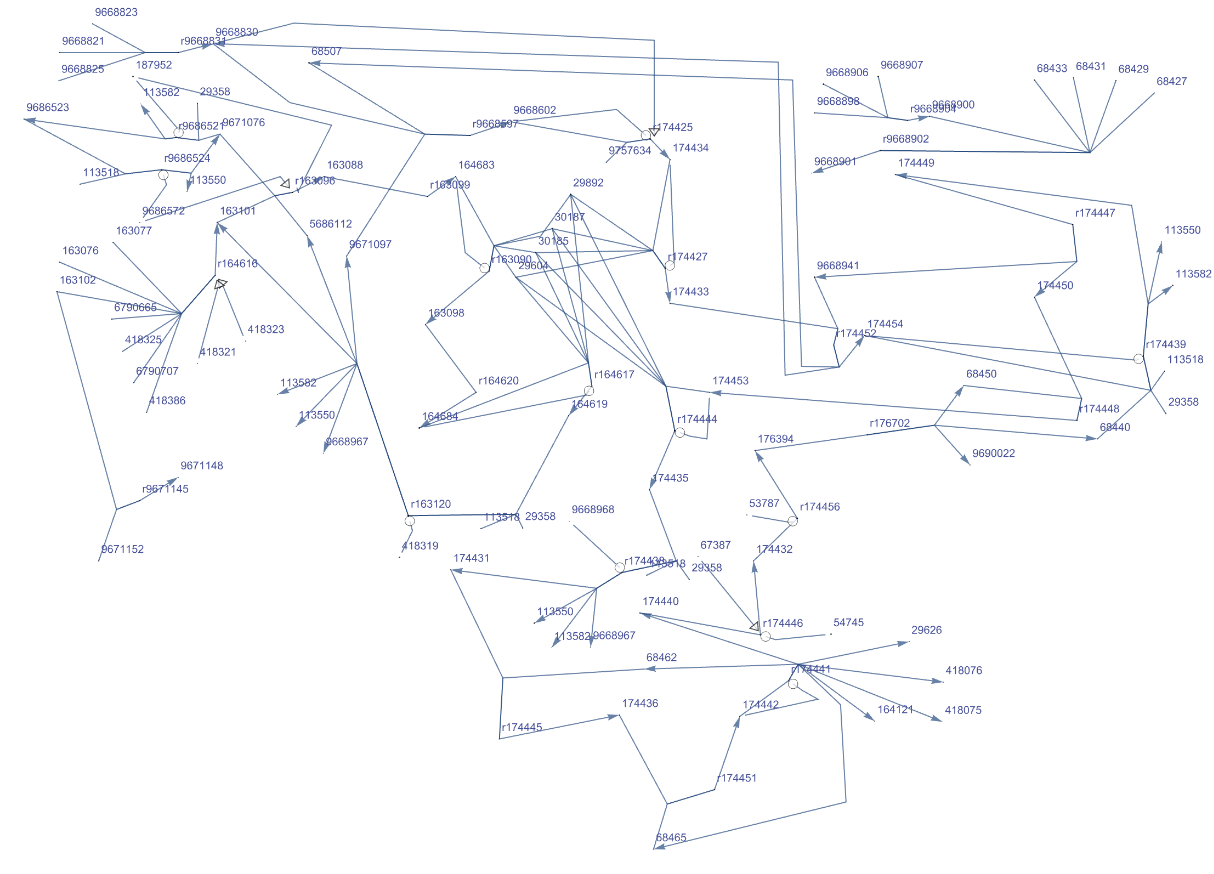 |
Get the Compartments of "R-HSA-180786" pathway for Homo sapiens:
| In[172]:= |
| Out[172]= |  |
Get the Glyph information to build the Graph for "R-HSA-180786" pathway for Homo sapiens:
| In[173]:= |
| Out[173]= | 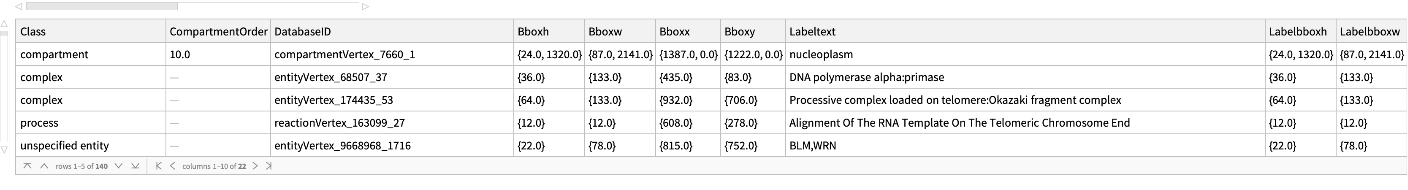 |
Get the Arc information to build the Graph for "R-HSA-180786" pathway for Homo sapiens:
| In[174]:= |
| Out[174]= | 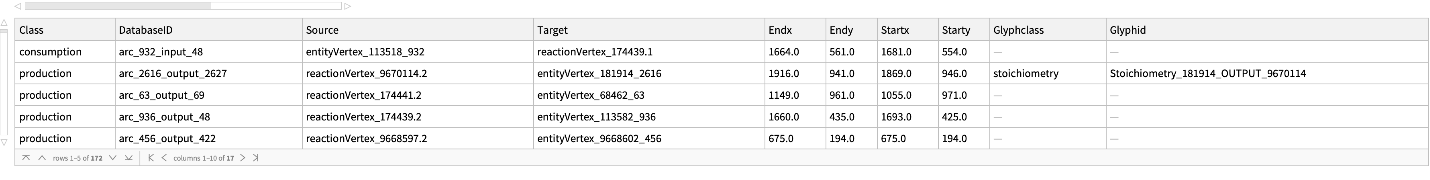 |
Get the Image of "R-HSA-180786" pathway for Homo sapiens:
| In[175]:= |
| Out[175]= | 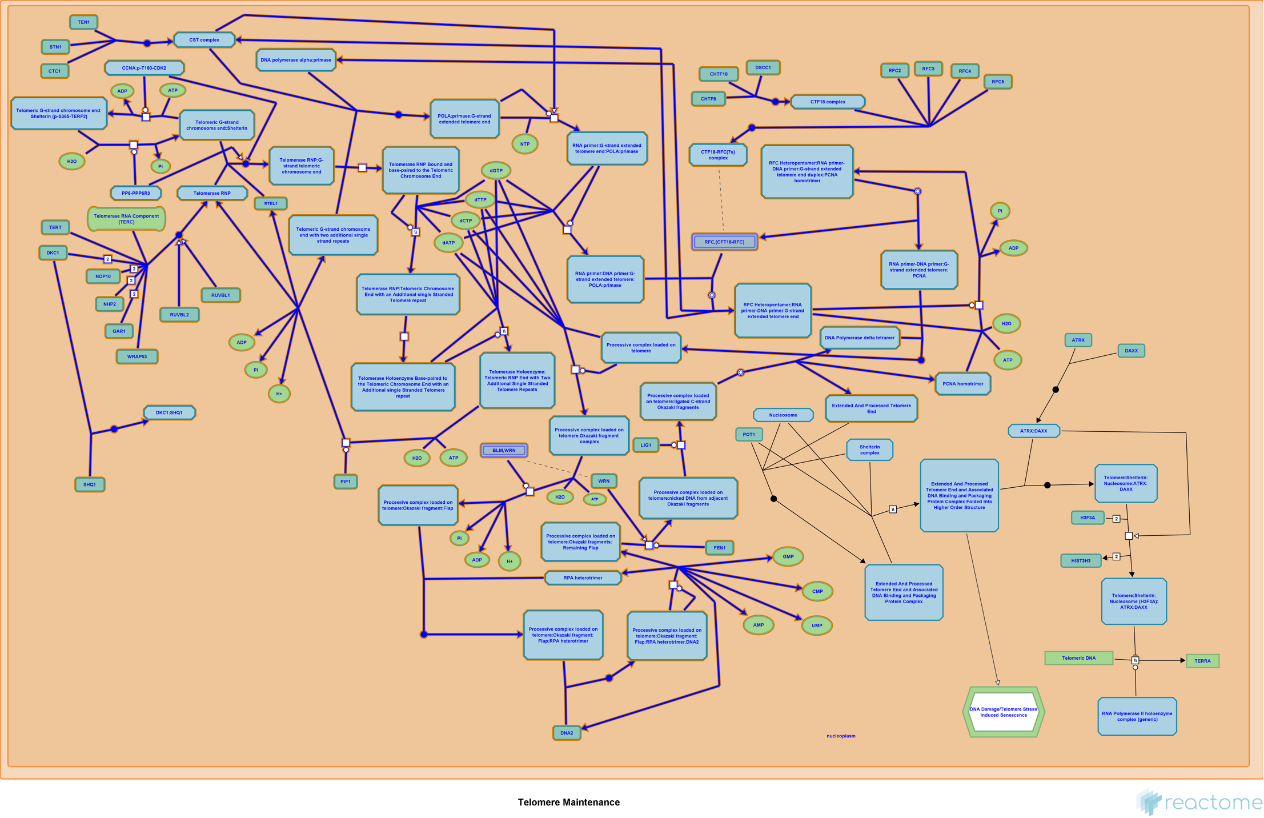 |
Get the ExternalIdentifiers of the entries in the "R-HSA-180786" pathway for Homo sapiens:
| In[176]:= |
| Out[176]= | 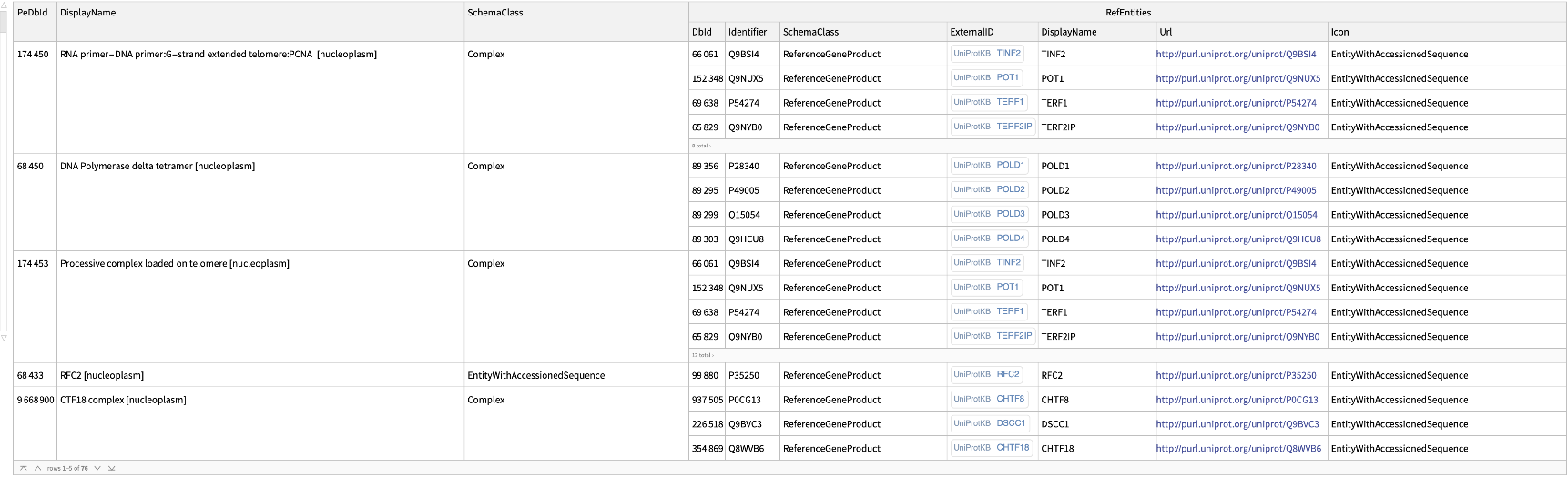 |
Customize the Graph of "R-HSA-180786" pathway for Homo sapiens:
| In[177]:= | ![ResourceFunction["ReactomePathways"]["R-HSA-180786", "Graph", VertexLabelStyle -> Directive[Black, 7],
VertexSize -> Scaled[.007], VertexStyle -> Table[i -> Red, {i, {"9668823", "9668821", "9668825"}}],
EdgeStyle -> Table[DirectedEdge[i, "9668831"] -> Red, {i, {"9668823", "9668821", "9668825"}}]]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/5a95dfc2d6879bad.png) |
| Out[177]= | 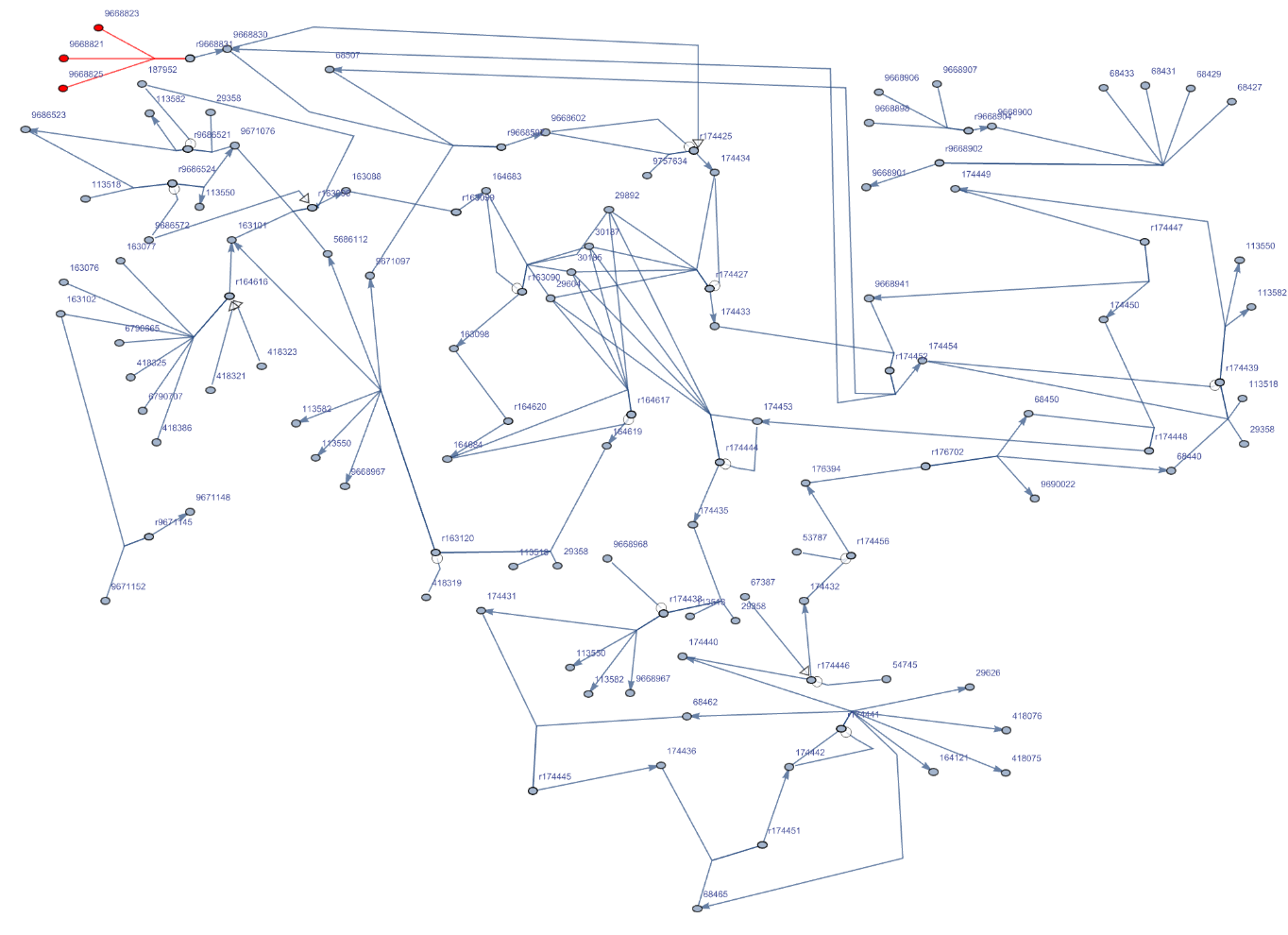 |
Delete the vertices representing reactions in the Graph of "R-HSA-180786" pathway for Homo sapiens:
| In[178]:= | ![ResourceFunction["ReactomePathways"]["R-HSA-180786", "Graph", "RemoveReactions" -> True, VertexLabelStyle -> Directive[Black, 7],
VertexSize -> Scaled[.007], VertexStyle -> Table[i -> Red, {i, {"9668823", "9668821", "9668825"}}],
EdgeStyle -> Table[DirectedEdge[i, "9668830"] -> Red, {i, {"9668823", "9668821", "9668825"}}]]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/3616625066bf43d0.png) |
| Out[178]= | 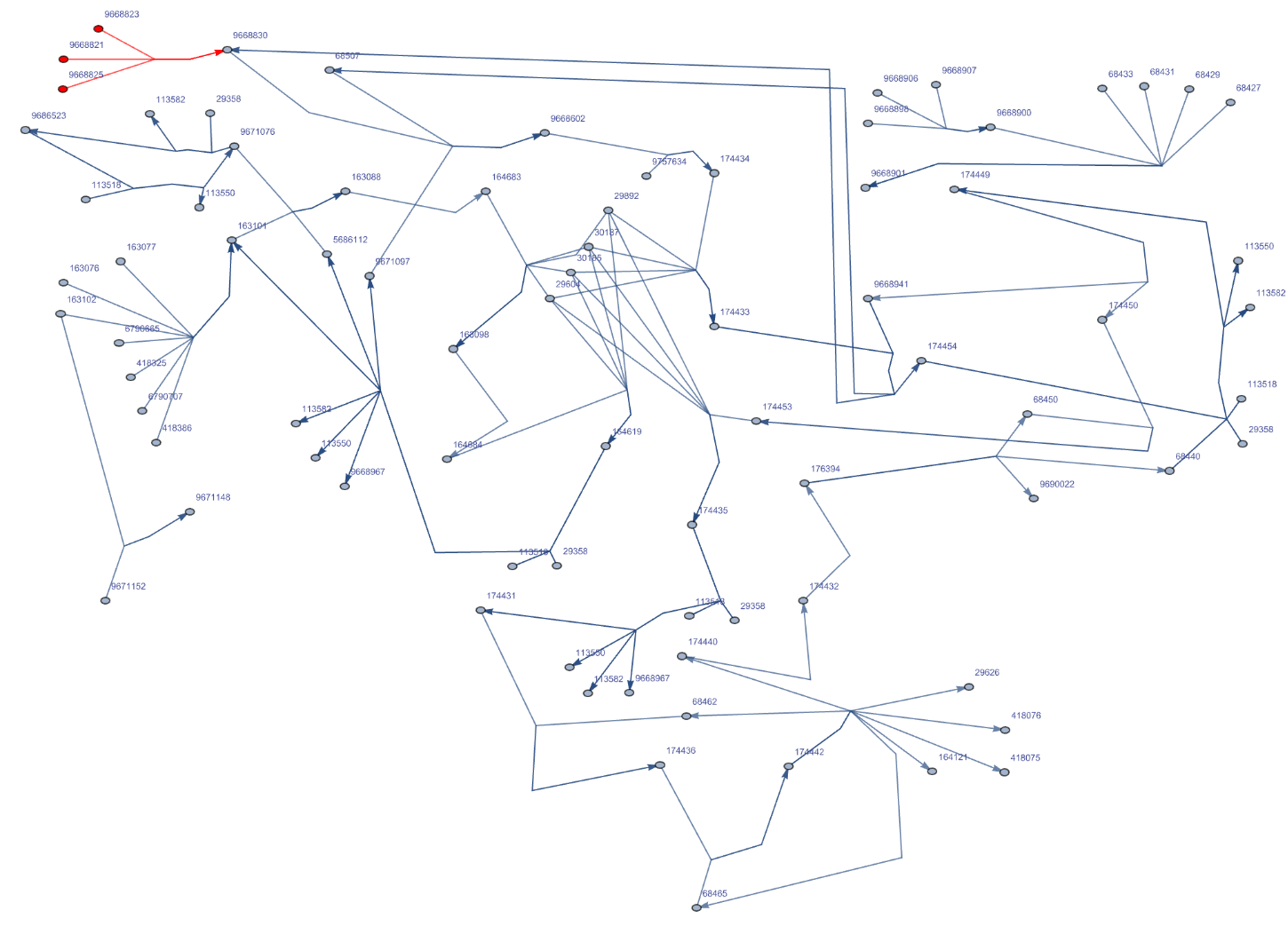 |
Sometimes is necessary to set the thickness of the edge style in order to display in a better way the edge highlighted:
| In[179]:= | ![ResourceFunction["ReactomePathways"]["R-HSA-174417", "Graph", "RemoveReactions" -> True, EdgeStyle -> ({"113518" \[DirectedEdge] "113550" -> RGBColor[
1, 0, 0]} /. Rule[a_, b_] :> Rule[a, Directive[Thick, b]]), VertexSize -> Medium, VertexLabelStyle -> Directive[Red, Italic, 4]]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/58ecca38fe770cd9.png) |
| Out[179]= | 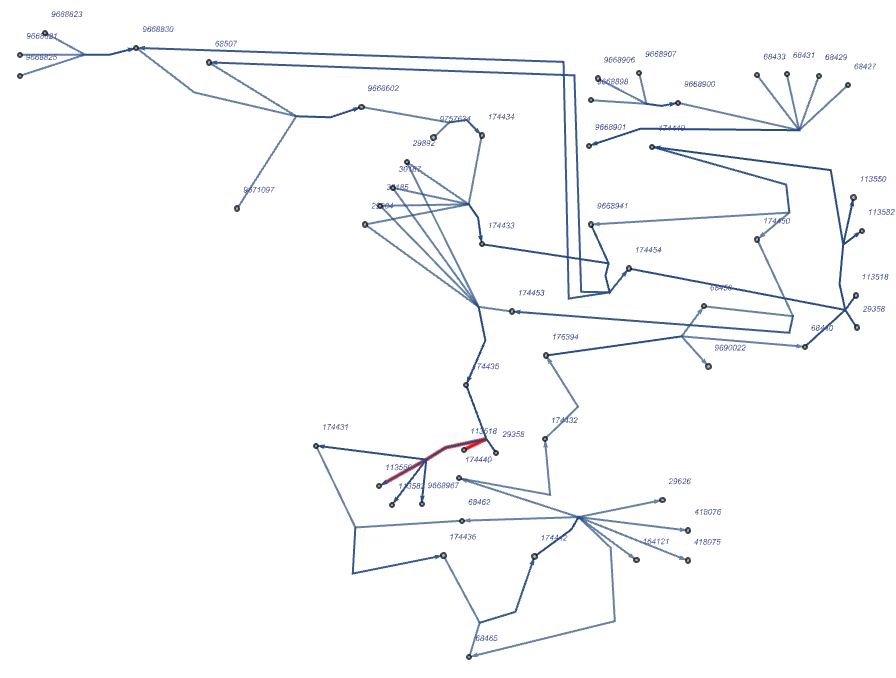 |
Customize the Image of the "R-HSA-180786" pathway for Homo sapiens:
| In[180]:= | ![ResourceFunction["ReactomePathways"]["R-HSA-180786", "Image",
"Extension" -> "png", "Quality" -> 10, "Flag" -> "R-HSA-174417", "FlagInteractors" -> "False", "Title" -> False, "Margin" -> 0, "Ehld" -> True, "DiagramProfile" -> "Modern", "Resource" -> "TOTAL", "AnalysisProfile" -> "Standard"]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/3c7bd47957fc8d52.png) |
| Out[200]= | 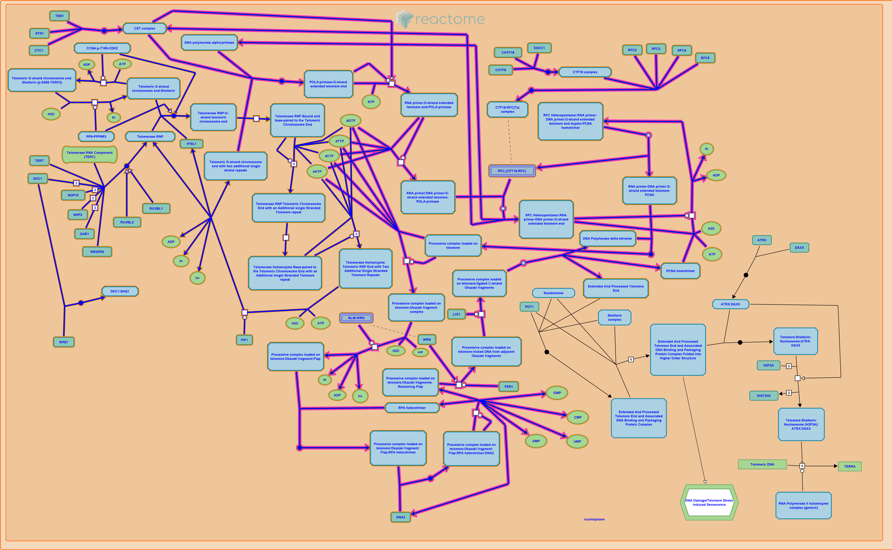 |
Look for pathways using keywords:
| In[201]:= |
| Out[202]= | 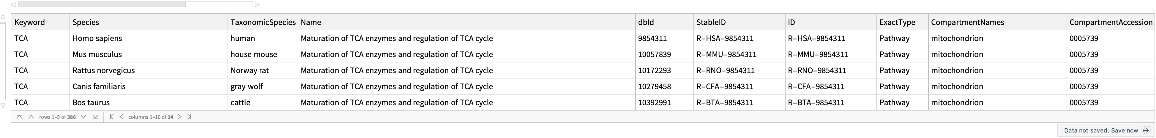 |
Look for pathways using keywords and for a particular species:
| In[203]:= |
| Out[203]= | 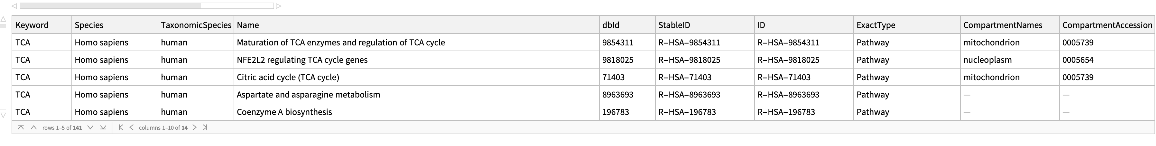 |
Pathways where an external identifier can be mapped to, using the name of the source and identifier:
| In[204]:= |
| Out[204]= | 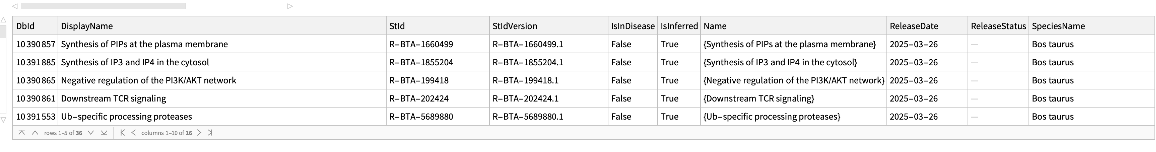 |
Pathways where an external identifier can be mapped to, using ExternalIdentifier:
| In[205]:= |
| Out[205]= | 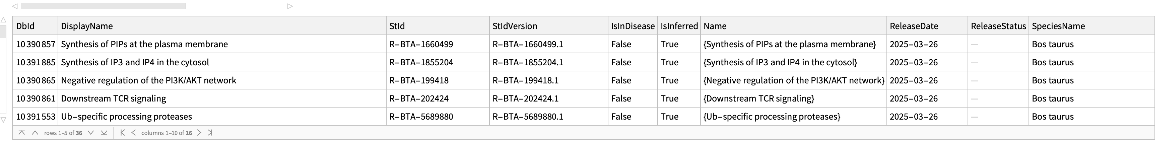 |
Pathways where an external identifier can be mapped to using the name of the source and identifier, specifying the species:
| In[206]:= | ![ResourceFunction[
"ReactomePathways"][{"UniProt", "PTEN"}, "QueryPaths", {"Species" -> Entity["TaxonomicSpecies", "HomoSapiens::4pydj"]}]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/50cf32ebd2e24f17.png) |
| Out[206]= | 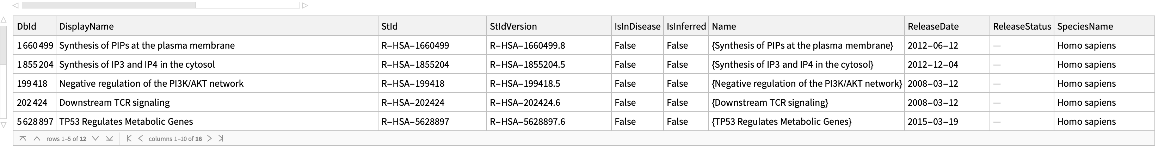 |
Pathways where an external identifier can be mapped to using ExternalIdentifier, specifying the species:
| In[207]:= | ![ResourceFunction["ReactomePathways"][
ExternalIdentifier["UniProtKBAccessionNumber", "PTEN"], "QueryPaths", {"Species" -> Entity["TaxonomicSpecies", "HomoSapiens::4pydj"]}]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/2cb62bbb66ab78c2.png) |
| Out[207]= | 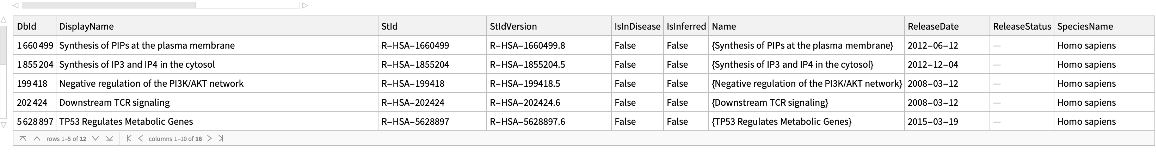 |
Reactions where an external identifier can be mapped to using ExternalIdentifier:
| In[208]:= |
| Out[208]= | 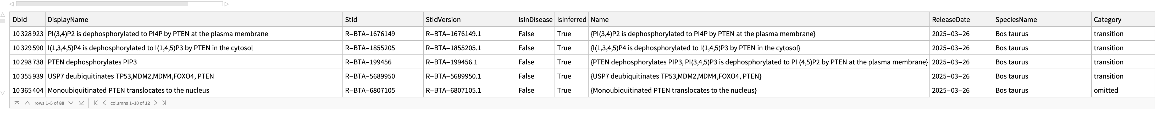 |
Reactions where an external identifier can be mapped to by specifying the species:
| In[209]:= | ![ResourceFunction["ReactomePathways"][
ExternalIdentifier["UniProtKBAccessionNumber", "PTEN"], "QueryReactions", {"Species" -> Entity["TaxonomicSpecies", "HomoSapiens::4pydj"]}]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/75cd310f8f509de9.png) |
| Out[209]= | 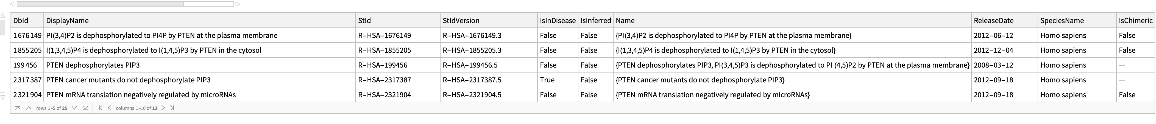 |
Get the vertex list of "R-HSA-180786" pathway for Homo sapiens:
| In[210]:= | ![vNames = Normal[ResourceFunction["ReactomePathways"]["R-HSA-180786", "Entries"][All, #DatabaseID -> #Name &]];
graph = ResourceFunction["ReactomePathways"]["R-HSA-180786", "Graph", "RemoveReactions" -> True];
sr = StringReplace[#, {"entityVertex" -> "species", "_" ~~ Shortest[x : DigitCharacter ..] ~~ EndOfString :> ""}] &;](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/6b519e627d10bdae.png) |
| In[211]:= |
| Out[211]= |
Get the edge list of "R-HSA-180786" pathway for Homo sapiens:
| In[212]:= |
| Out[212]= |
Get the number of vertices for "R-HSA-180786" pathway for Homo sapiens:
| In[213]:= |
| Out[213]= |
Get the number of edges of "R-HSA-180786" pathway for Homo sapiens:
| In[214]:= |
| Out[214]= |
Get the vertex degrees:
| In[215]:= |
| Out[215]= |
Make a histogram:
| In[216]:= |
| Out[216]= | 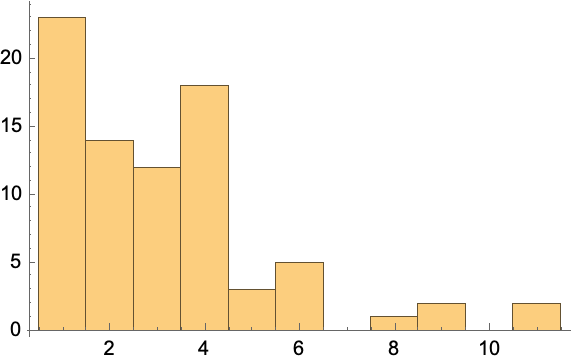 |
Get the vertex out-degree list:
| In[217]:= |
| Out[217]= |
Make a histogram:
| In[218]:= |
| Out[219]= | 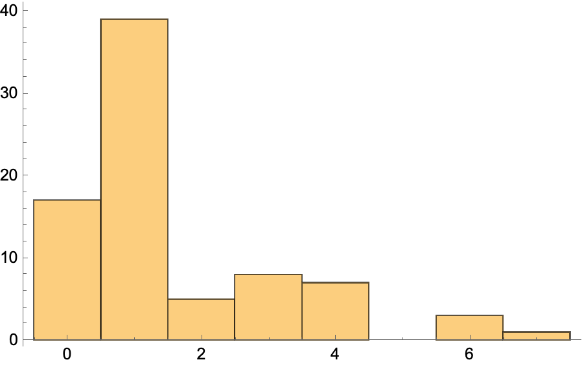 |
Get the vertex in-degree list:
| In[220]:= |
| Out[220]= |
Make a histogram:
| In[221]:= |
| Out[221]= | 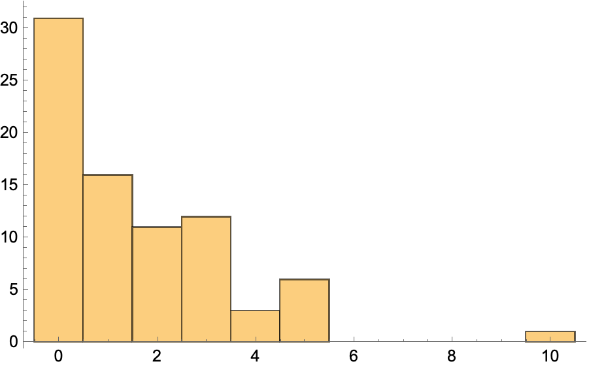 |
Get the BetweennessCentrality:
| In[222]:= |
| Out[222]= |
Make a histogram:
| In[223]:= |
| Out[223]= |  |
You will get $Failed if the pathway id is incorrect:
| In[224]:= |
| Out[224]= |
Highlight the sub pathways in the "R-HSA-180786" pathway for Homo sapiens:
| In[225]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/3223e55a-d5a5-4565-98a2-fa4f29edf05e"]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/52a4130dbe99f768.png) |
| In[226]:= | ![Manipulate[
edgeList2 = EdgeList[lowLevelsPathsGraphs[path]];
edgeListFinal = If[
Length[edgeList2] == Length[Intersection[edgeList1, edgeList2]],
Thread[edgeList2 -> Red],
SortBy[#, Last][[-1]][[1, 2]] & /@ Table[{i, j -> Red} -> SmithWatermanSimilarity[ToString@i, ToString@j], {i, edgeList2}, {j, edgeList1}]
];
ResourceFunction["ReactomePathways"]["R-HSA-180786", "Graph", "RemoveReactions" -> True, EdgeStyle -> edgeListFinal, VertexSize -> Medium, VertexLabelStyle -> Directive[Red, Italic, 4],
PlotLabel -> Style["Extension of telomeres \[OpenCurlyDoubleQuote]R-HSA-180786\[CloseCurlyDoubleQuote] pathway", 10, Black]] , {path, Keys[lowLevelsPathsGraphs]}
]](https://www.wolframcloud.com/obj/resourcesystem/images/ce8/ce875dfd-6780-4ce9-bb63-bdfffa1bd091/793de926b54d4379.png) |
| Out[226]= | 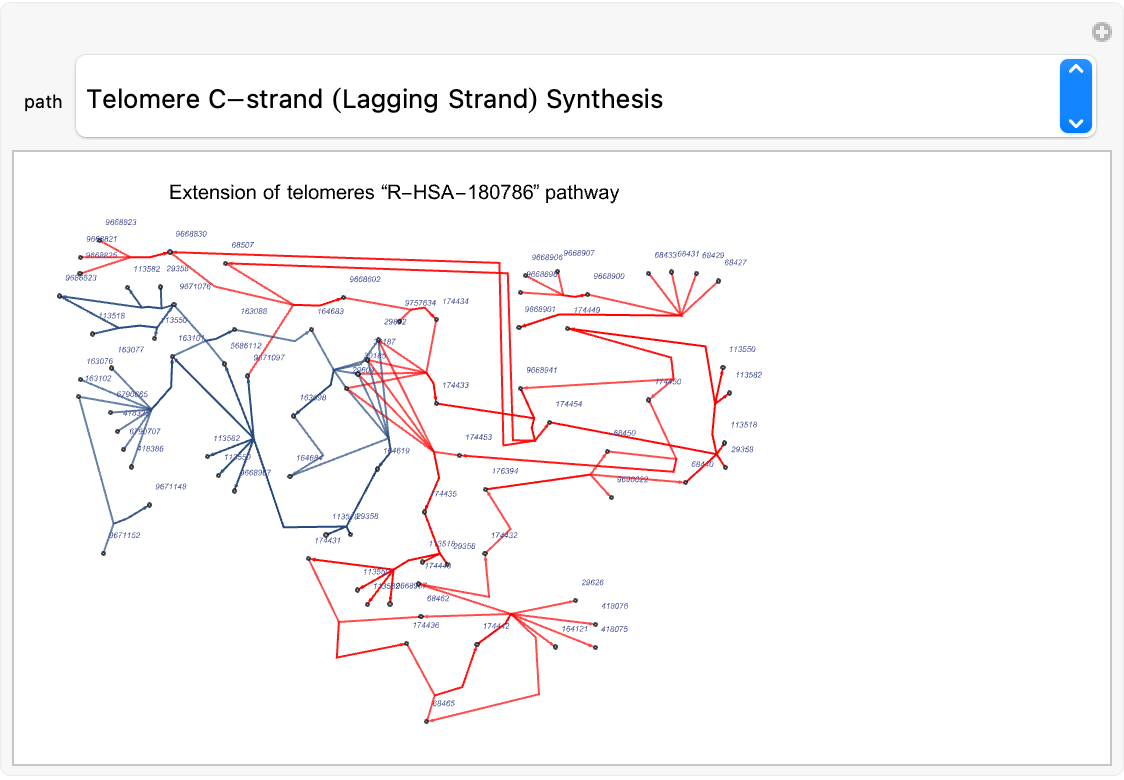 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License