Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get information about a KEGG orthology
ResourceFunction["KEGGOrthology"]["OrthologyEntries"] gives the Dataset with basic information for all KEGG orthologies. | |
ResourceFunction["KEGGOrthology"][keggcode,prop] gives a property prop for a specific keggcode. | |
ResourceFunction["KEGGOrthology"][keyword,"Query"] gives a Dataset for a specific keyword referring to an orthology. |
| "Orthology" | information for keggcode referring to KEGG orthology (i.e., K00001) |
| "Entry" | ExternalIdentifier of the orthology KEGG code |
| "EntryType" | the orthology type |
| "Symbol" | the orthology name |
| "FullName" | the orthology full name |
| "EntryInName" | the enzyme specified in the orthology name |
| "Reaction" | reactions involving the orthology |
| "PathwayExternalID" | ExternalIdentifier of the pathways involving the orthology |
| "PathwayHyperlink" | hyperlink of the pathways involving the orthology |
| "ModuleExternalID" | ExternalIdentifier of the modules involving the orthology |
| "ModuleHyperlink" | hyperlink of the modules involving the orthology |
| "Disease" | diseases involving the orthology |
| "Brite" | orthology classification |
| "OtherDBs" | lists to other classification systems |
| "Genes" | genes in KEGG organisms that belong to the orthology group |
| "Reference" | reference information |
Get the list of all the orthology entries in the KEGG orthology database:
| In[1]:= |
| Out[2]= | 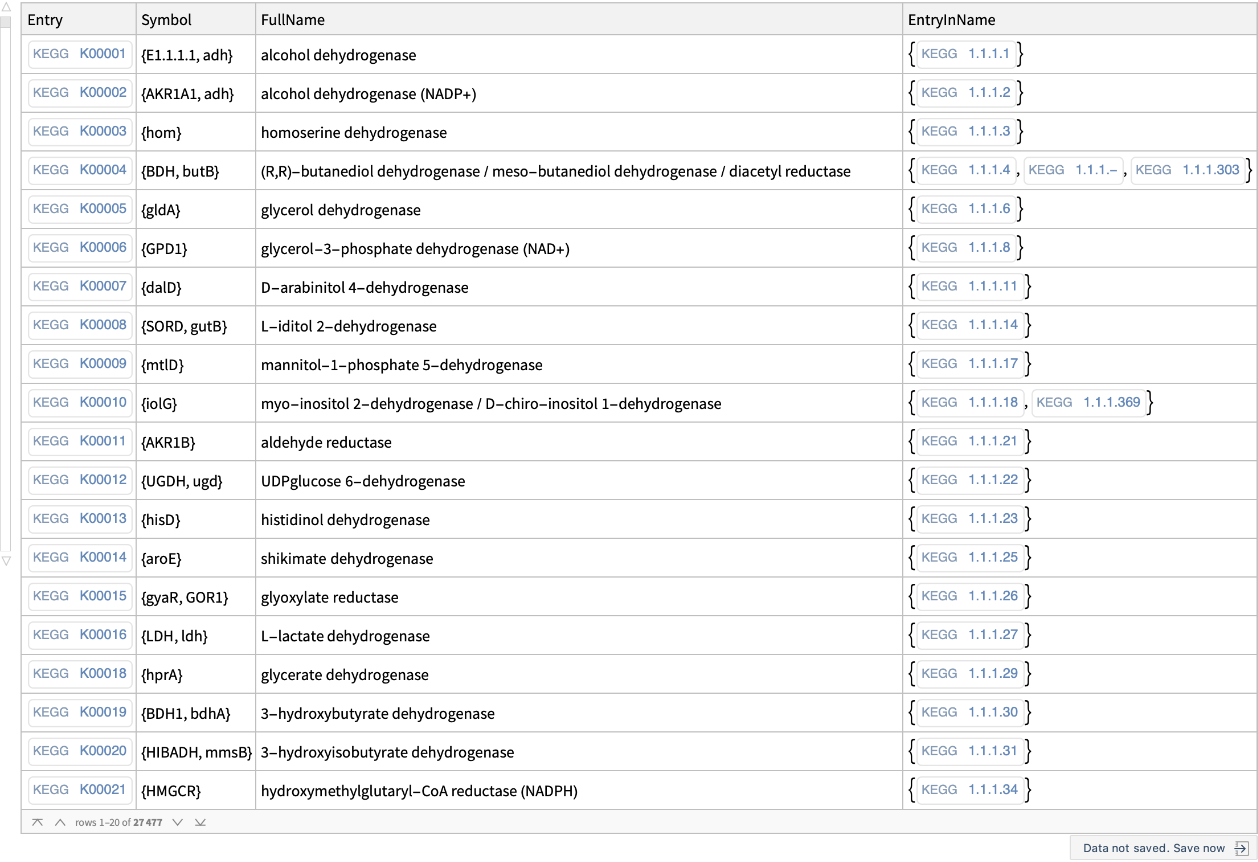 |
Get information from KEGG about a specific orthology:
| In[3]:= |
| Out[3]= | 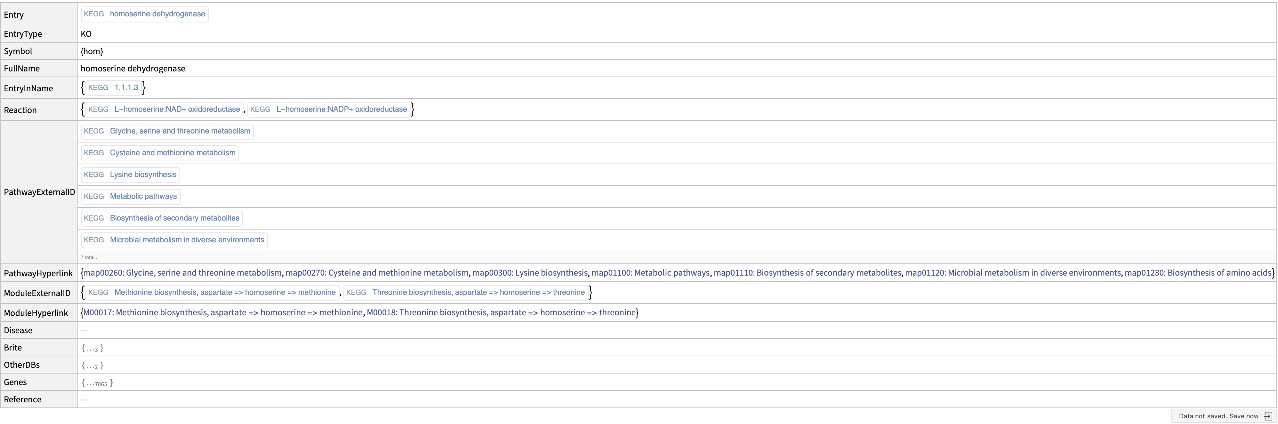 |
Get information from KEGG about a list of orthology entries:
| In[4]:= |
| Out[4]= |  |
Get the KEGG orthology codes for a specific query:
| In[5]:= |
| Out[5]= | 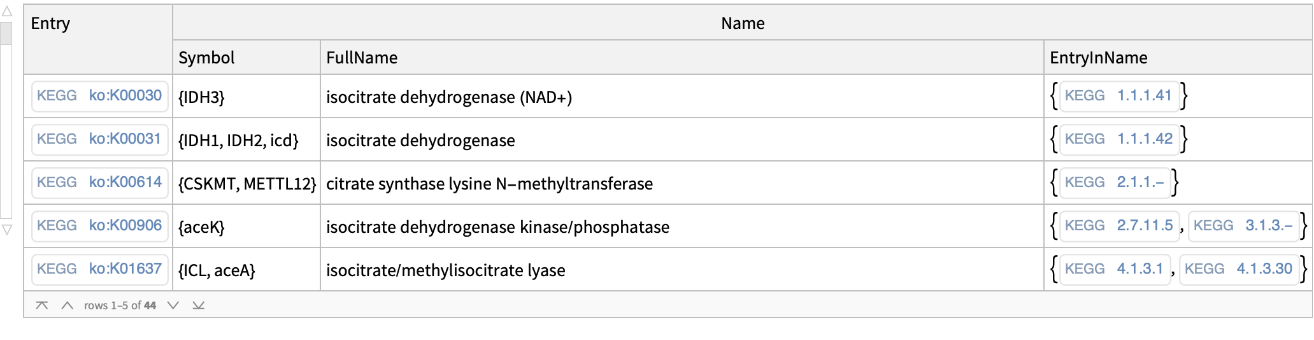 |
Keywords can have more than one part:
| In[6]:= |
| Out[6]= | 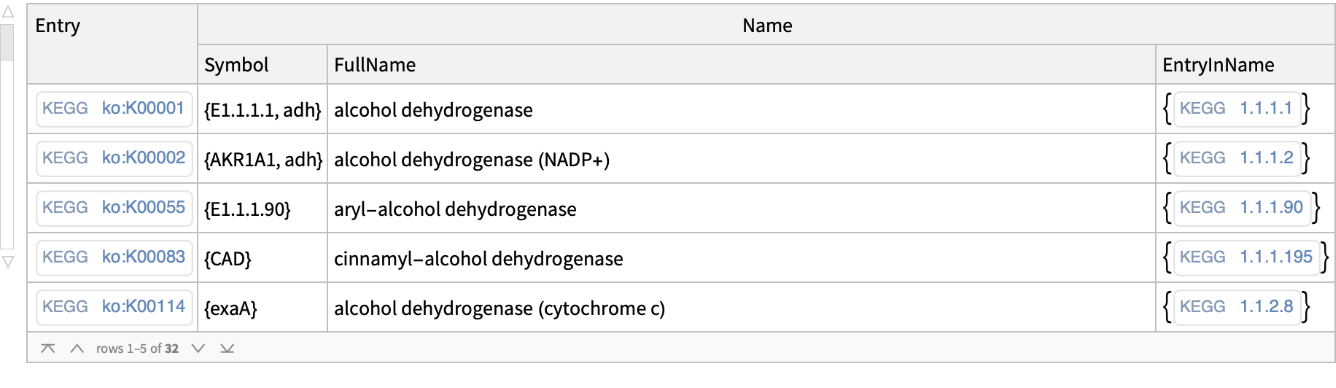 |
Get the dataset properties independently for each module:
| In[7]:= |
| Out[7]= |  |
Get the dataset properties independently for a list of modules:
| In[8]:= |
| Out[8]= |  |
Another way to get the information from KEGG about a specific orthology:
| In[9]:= |
| Out[9]= |  |
Another way to get the information from KEGG about a specific list of orthology entries:
| In[10]:= |
| Out[10]= |
Only known keywords are supported:
| In[11]:= |
| Out[11]= |
Get the pathway and reactions associated with the K0003 orthology:
| In[12]:= |
| Out[12]= |  |
| In[13]:= |
| Out[13]= |
The next are the compounds involved in the previous reactions:
| In[14]:= |
| In[15]:= |
Select one species that contains the orthology K00003:
| In[16]:= |
| Out[16]= |
Get the list of edges for the biosynthesis of amino acids, one of the pathways associated with the K00003 orthology:
| In[17]:= | ![vNamesRules = Normal[ResourceFunction[
ResourceObject[<|"Name" -> "KEGGPathway", "ShortName" -> "KEGGPathway", "UUID" -> "c26fb78f-9391-4e5b-8348-1b3a9bcb8293", "ResourceType" -> "Function", "Version" -> "1.2.1", "Description" -> "Get the graph and additional information of a KEGG pathway", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$cf8afda6cda14c308cd1e656f8629ece`KEGGPathway", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/81066f03-423e-4566-800b-da9d03968a36"]|>, {ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"}]]["mpha", "01230", "Entries"][
All, #EntryID -> #EntryGraphName &]];
graph = ResourceFunction[
ResourceObject[<|"Name" -> "KEGGPathway", "ShortName" -> "KEGGPathway", "UUID" -> "c26fb78f-9391-4e5b-8348-1b3a9bcb8293", "ResourceType" -> "Function", "Version" -> "1.2.1", "Description" -> "Get the graph and additional information of a KEGG pathway", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$cf8afda6cda14c308cd1e656f8629ece`KEGGPathway", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/81066f03-423e-4566-800b-da9d03968a36"]|>, {ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"}]]["mpha", "01230", "Graph"];](https://www.wolframcloud.com/obj/resourcesystem/images/c40/c40bba05-c70e-4b41-b65d-276f55f97aec/7cf9ceb6864d8649.png) |
| In[18]:= |
| Out[18]= |  |
Using the information about reactions and compounds, we get the list of edges and vertices to highlight the reaction in biosynthesis of amino acids pathway that is catalyzed by the K00003 orthology:
| In[19]:= |
| Out[19]= |
| In[20]:= |
| Out[20]= |
Highlighting in red (i.e., vertices and edges) the reaction catalyzed by the K00003 orthology:
| In[21]:= | ![ResourceFunction[
ResourceObject[<|"Name" -> "KEGGPathway", "ShortName" -> "KEGGPathway", "UUID" -> "c26fb78f-9391-4e5b-8348-1b3a9bcb8293", "ResourceType" -> "Function", "Version" -> "1.2.1", "Description" -> "Get the graph and additional information of a KEGG pathway", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$cf8afda6cda14c308cd1e656f8629ece`KEGGPathway", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/81066f03-423e-4566-800b-da9d03968a36"]|>, {ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"}]]["mpha", "01230", "Graph",
VertexSize -> Medium,
VertexStyle -> vertx,
EdgeStyle -> edges]](https://www.wolframcloud.com/obj/resourcesystem/images/c40/c40bba05-c70e-4b41-b65d-276f55f97aec/773480cea72acdd0.png) |
| Out[21]= | 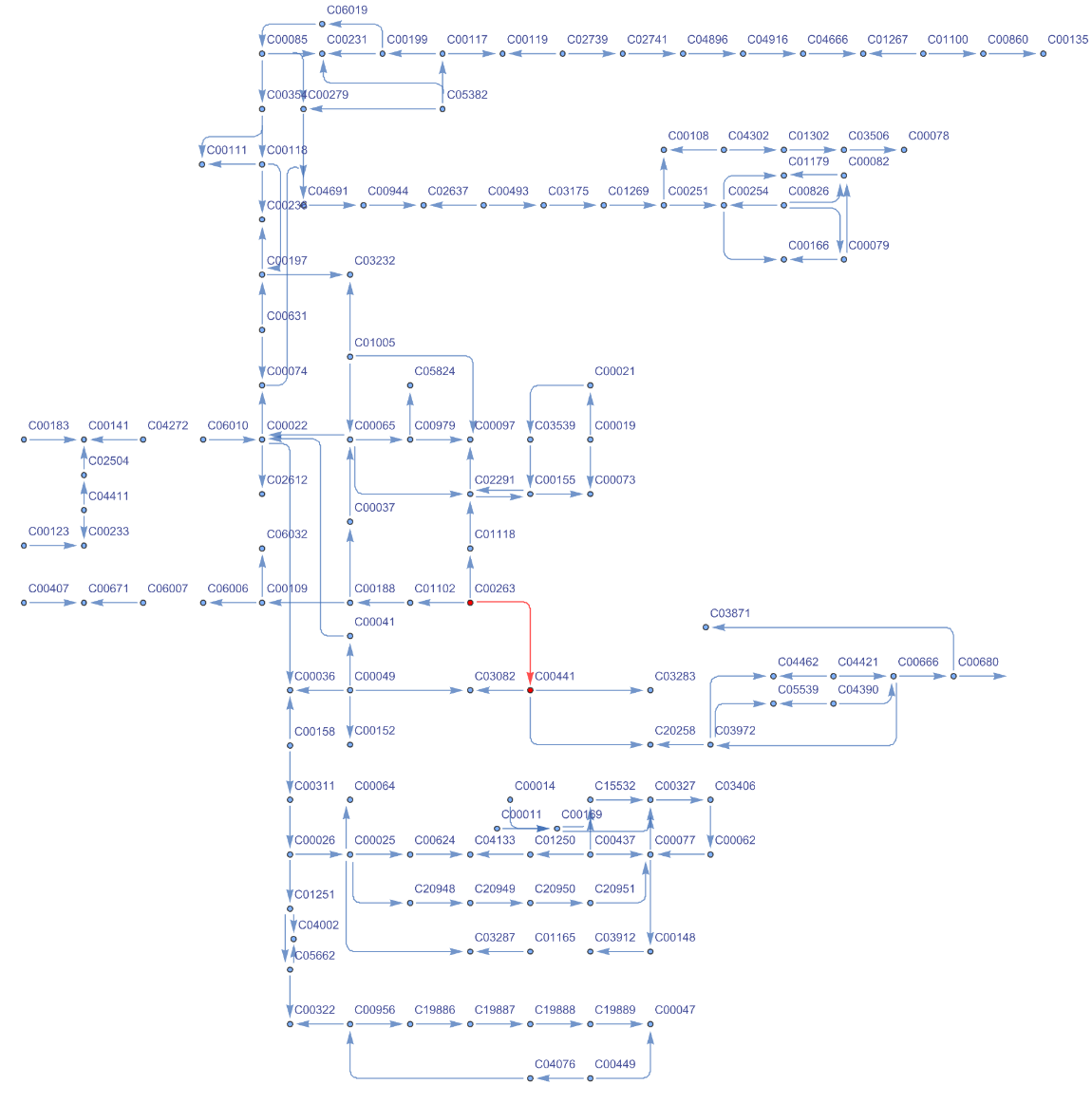 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License