Basic Examples (5)
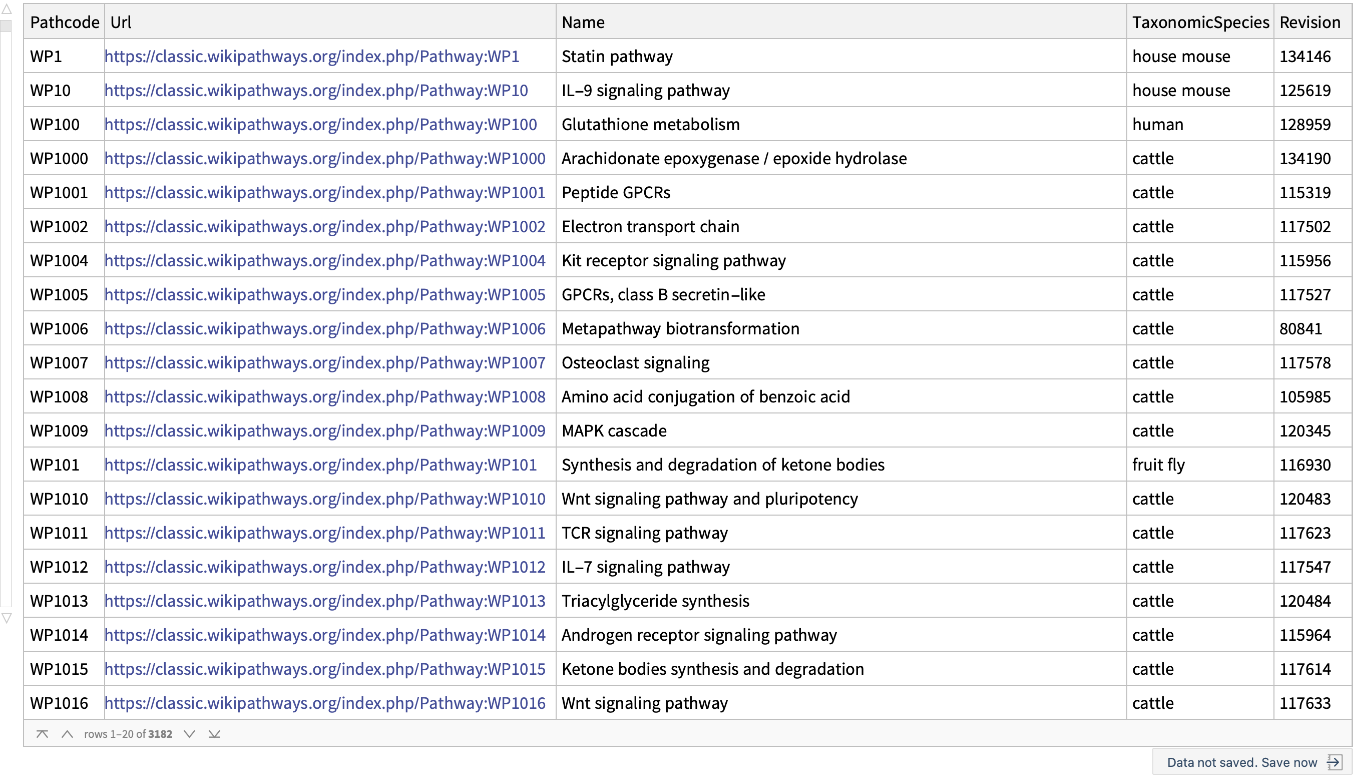
Get basic information for all the WikiPathways including their revision number:
Get information for a specific WikiPathway:
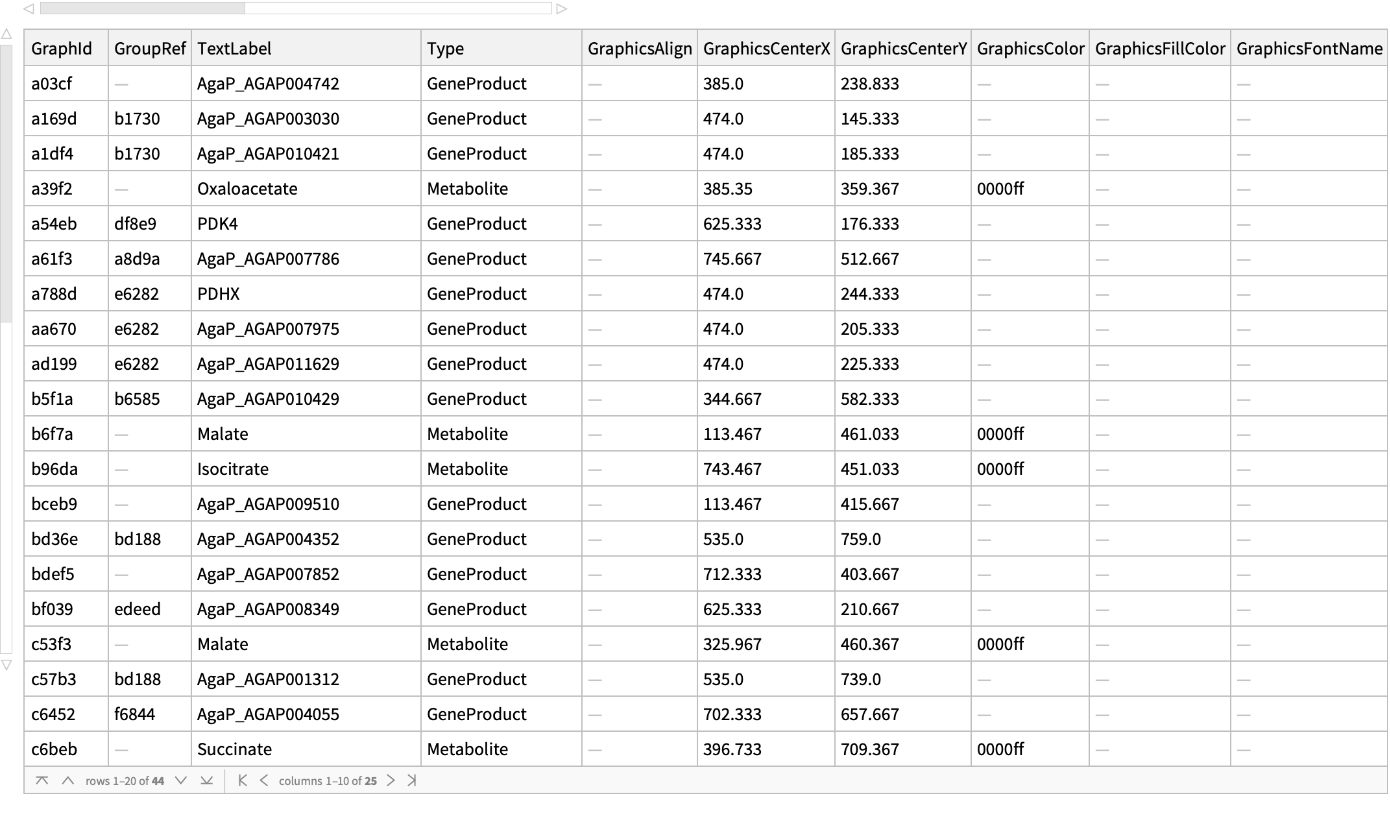
Get all the information related to nodes:
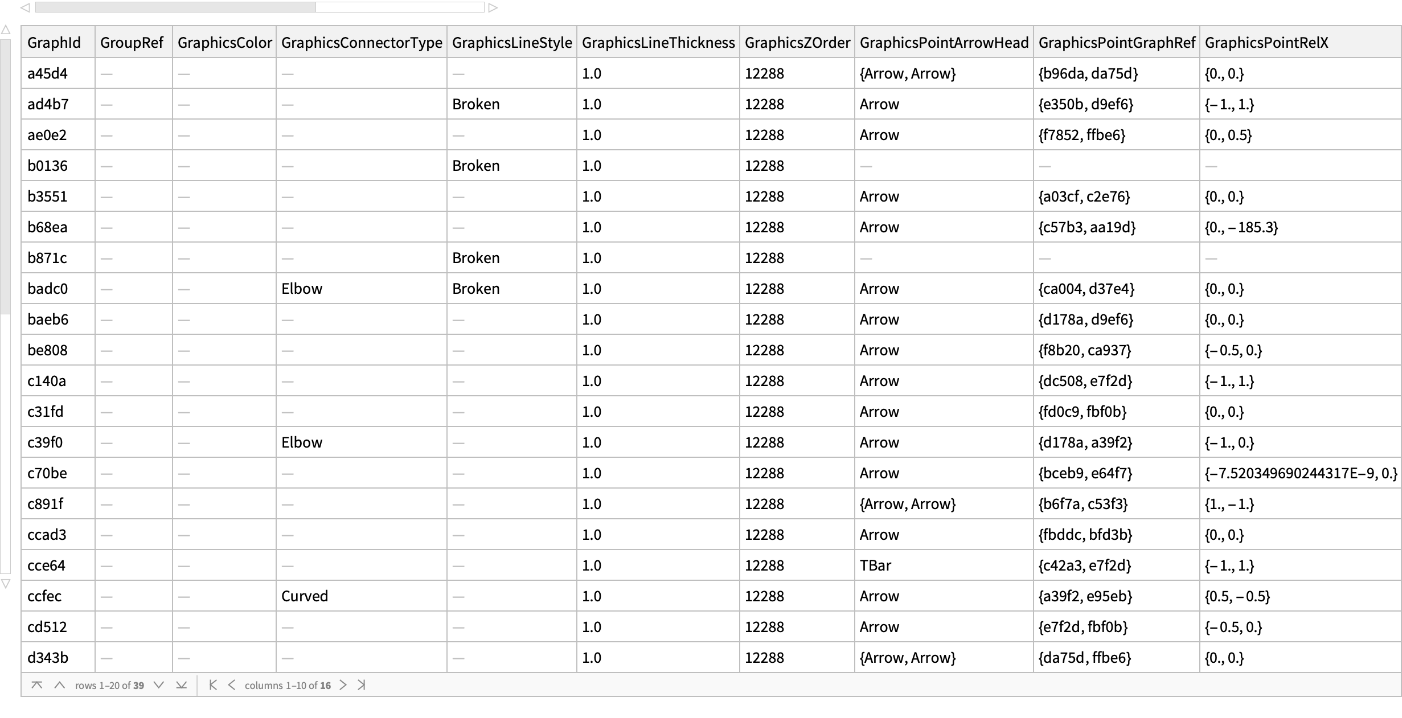
Get all the information related to interactions or lines connecting elements of the graphics with a biological meaning:
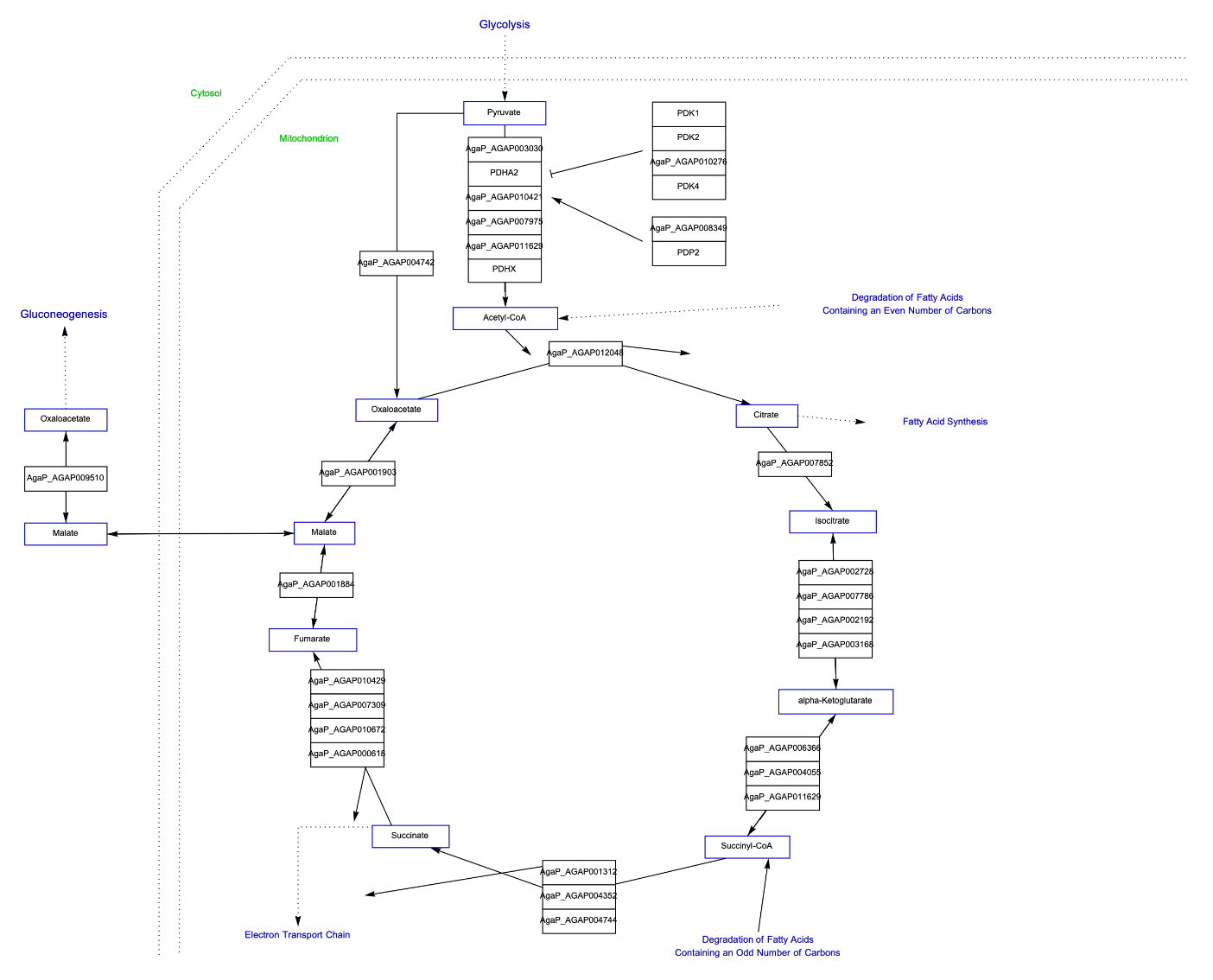
Get the Graphics of "TCA cycle" for African malaria mosquito. Get the complete names of elements and additional information of edges hovering over them. Click the polygons to get a popup window with links to more information:
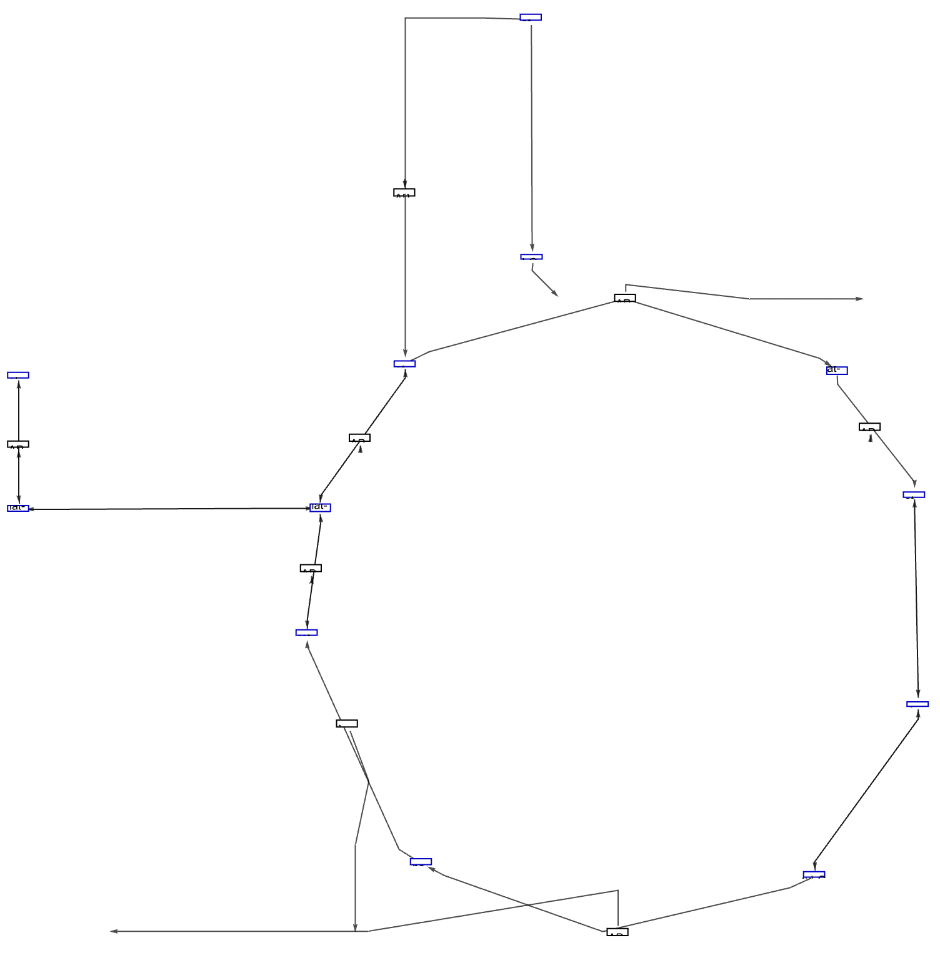
Get the Graph version of the Graphics of "TCA cycle" for African malaria mosquito. Get the complete names of elements and additional information of edges hovering over them. Click the polygons to get a popup window with links to more information:
Scope (3)
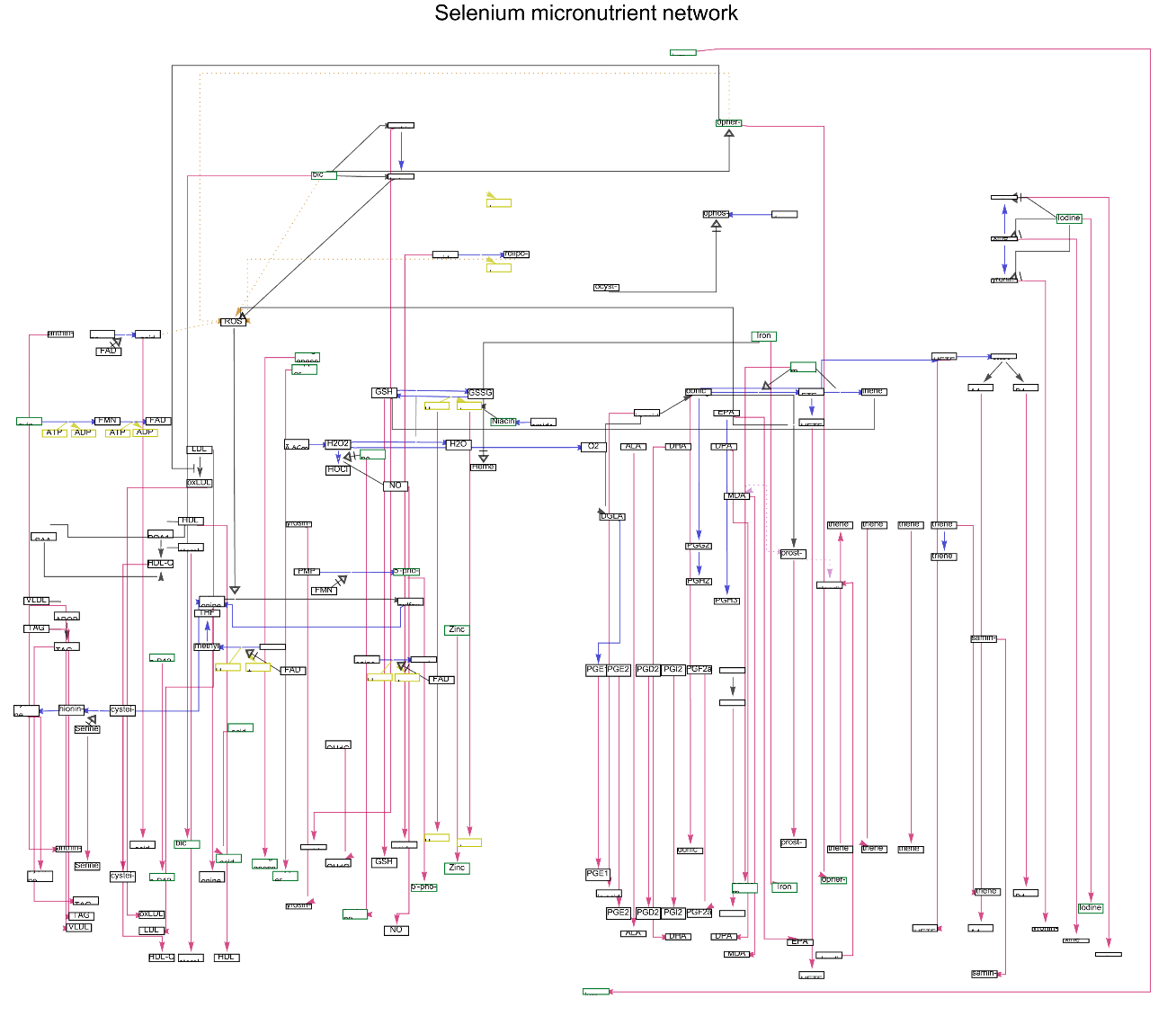
Get all the GraphicsPointArrowHead types in "Selenium micronutrient network":
Select the "mim-inhibition" type of GraphicsPointArrowHead from "Selenium micronutrient network" Graph:
Get all the Types of nodes in "Selenium micronutrient network":
Select the "Metabolite" Type of node from "Selenium micronutrient network" Graph:
Options (4)
Customize the visualization of "TCA cycle" Graph for African malaria mosquito:
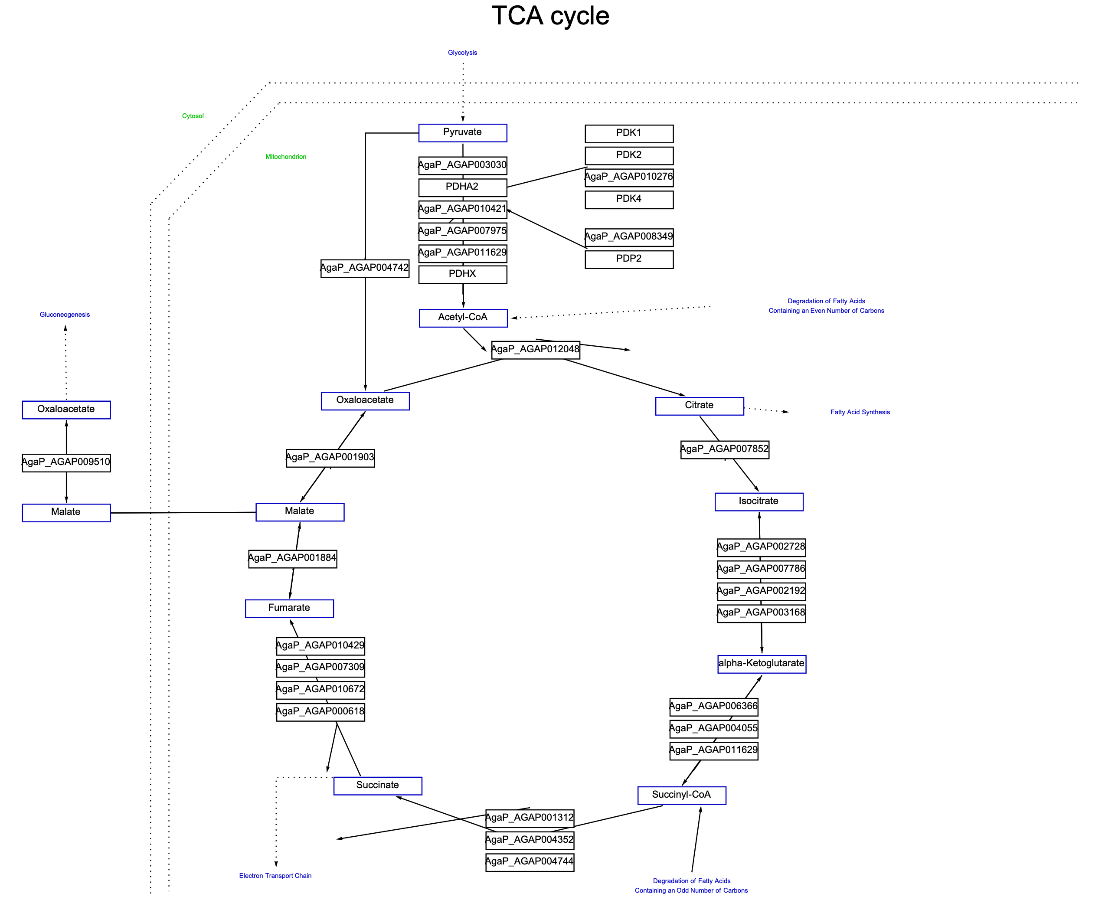
Customize the visualization of "TCA cycle" Graphics for African malaria mosquito:
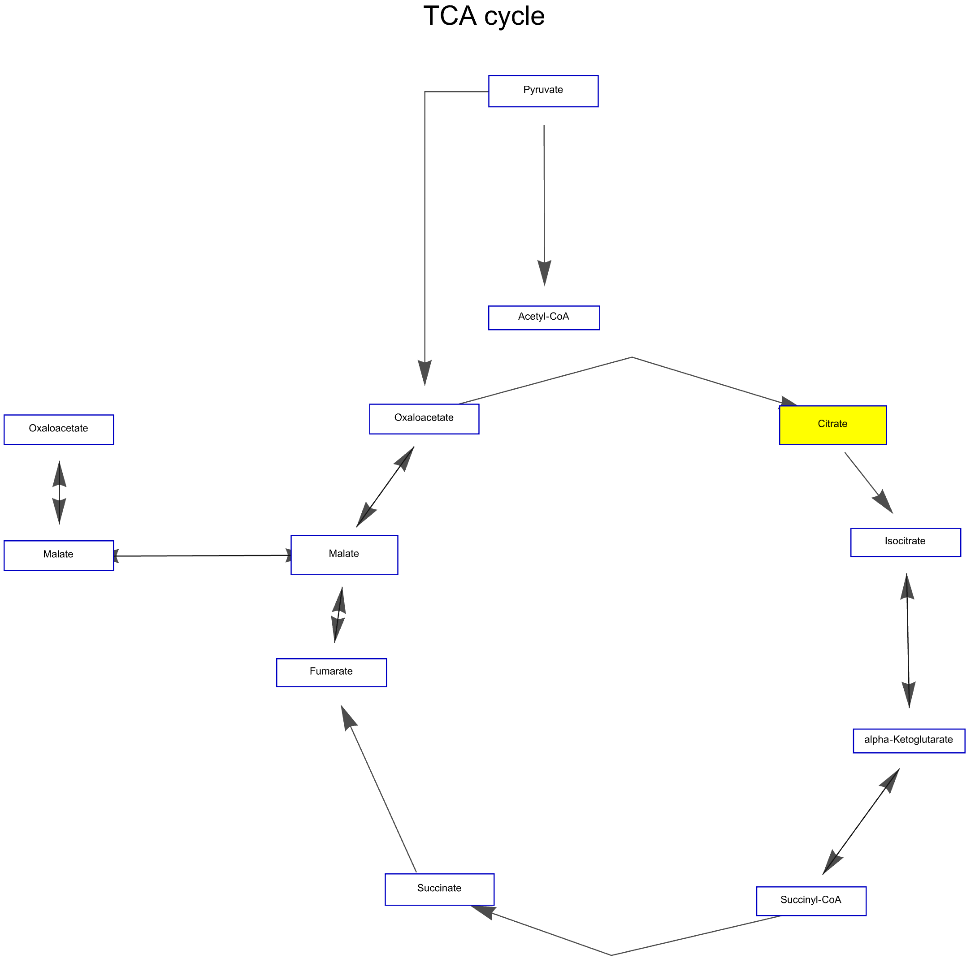
Highlight nodes of the "TCA cycle" Graph for African malaria mosquito:
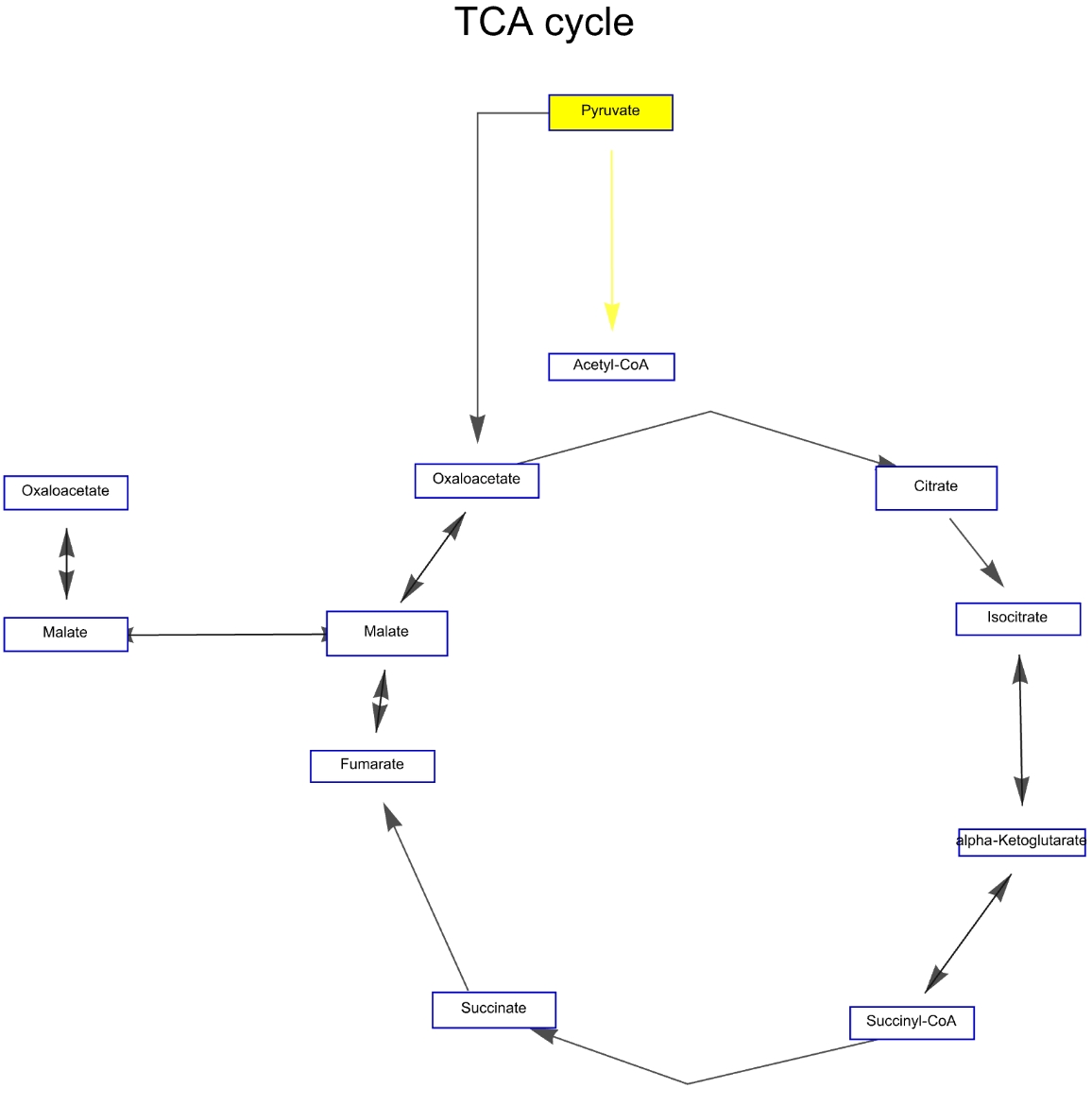
Highlight edges of the "TCA cycle" Graph for African malaria mosquito:
Properties and Relations (8)
Get the vertex list:
Get the edges list:
Get the number of vertex:
Get the number of edges:
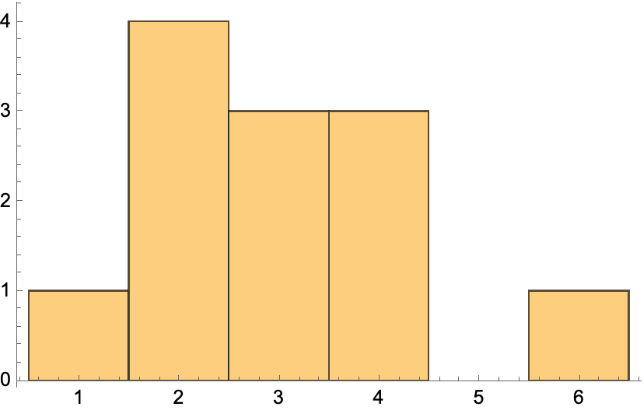
Get the vertex degree and histogram:
Get the vertex out degree list and histogram:
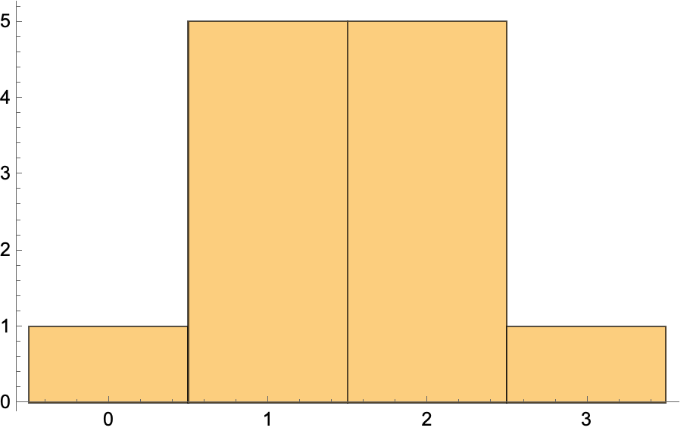
Get the vertex in degree list and histogram:
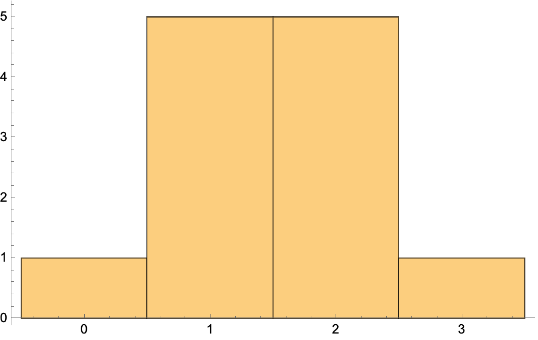
Get the BetweennessCentrality and histogram:
Possible Issues (3)
Issue could be found if the pathway id is incorrect:
Issue could be found if the property in misspelled:
Issue could be found if the graph data is missing:
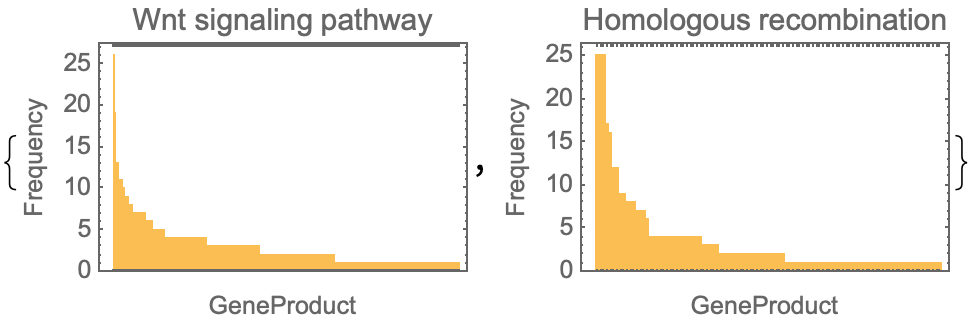
Neat Examples (1)
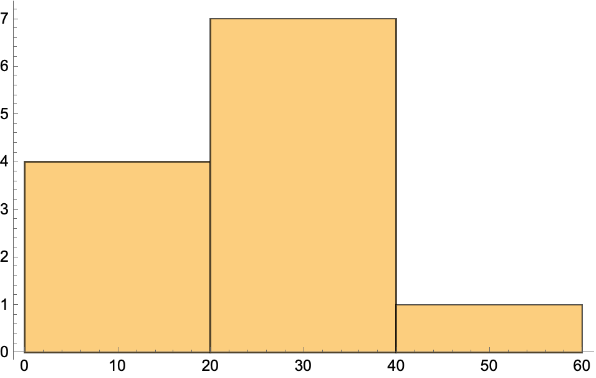
Get the gene products frequencies of the same pathway in different species:

![arrowheads = DeleteDuplicates[
Normal[ResourceFunction["WikiPathways"]["WP3257", "Interaction"][
All, #GraphicsPointArrowHead &]]]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/2018a2d8e8b59cc2.png)
![ResourceFunction["WikiPathways"]["WP3257", "Graph", "InteractionType" -> {"mim-inhibition"}, PlotLabel -> Style["Selenium micronutrient network", Black, Large]]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/00b4ed85c297f8fd.png)
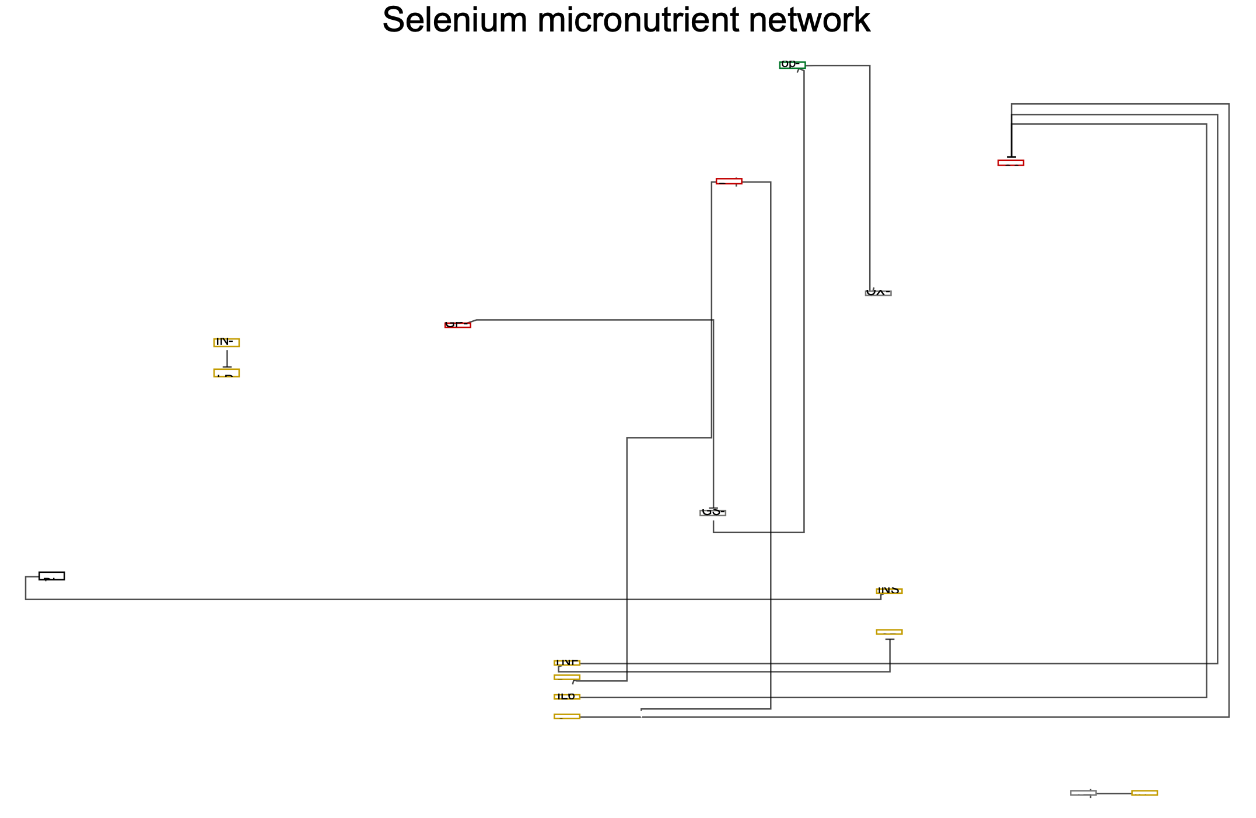
![ResourceFunction["WikiPathways"]["WP3257", "Graph", "DataNodeType" -> {"Metabolite"}, PlotLabel -> Style["Selenium micronutrient network", Black, Large]]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/7b2b37c063173dbb.png)
![ResourceFunction["WikiPathways"]["WP1227", "Graph", "ArrowHeadSize" -> 0.03, "Distance" -> 0.03, "DeleteAnchors" -> True,
VertexSize -> 1, PlotLabel -> Style["TCA cycle", Black, Large]]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/4b4a8783f2a5e53c.png)
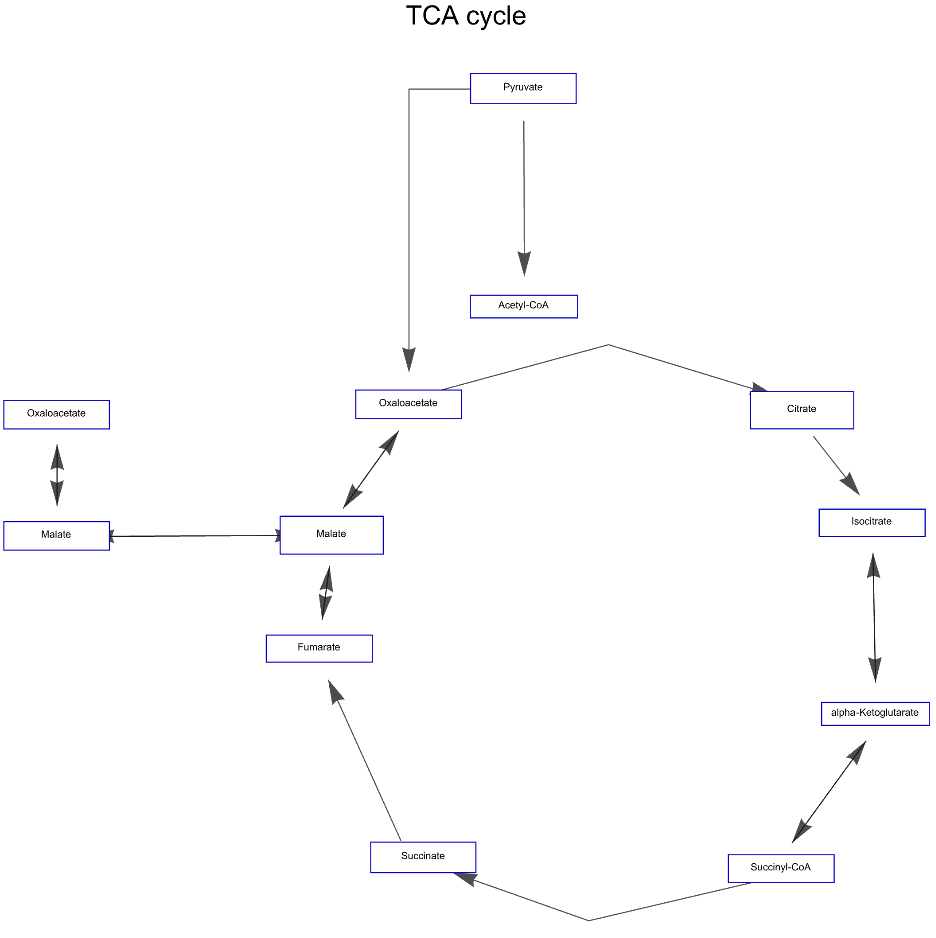
![ResourceFunction["WikiPathways"]["WP1227", "Graphics", "ArrowHeadSize" -> 0.006, "LabelSize" -> 6, "NodeHeight" -> 16, "NodeWidth" -> 80, PlotLabel -> Style["TCA cycle", Black, Large]]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/2da0272d834341b8.png)
![ResourceFunction["WikiPathways"]["WP1227", "Graph", "ArrowHeadSize" -> 0.03, "Distance" -> 0.03, "DeleteAnchors" -> True,
VertexSize -> 1, PlotLabel -> Style["TCA cycle", Black, Large], VertexStyle -> {"Citrate" -> Yellow}]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/0cd5fa0898fb10b3.png)
![ResourceFunction["WikiPathways"]["WP1227", "Graph", "ArrowHeadSize" -> 0.03, "Distance" -> 0.03, "DeleteAnchors" -> True,
VertexSize -> 1, PlotLabel -> Style["TCA cycle", Black, Large], VertexStyle -> {"Pyruvate" -> Yellow}, EdgeStyle -> {"Pyruvate" -> "Acetyl-CoA" -> Yellow}]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/5cce3c4527aec01a.png)
![vNames = Normal[ResourceFunction["WikiPathways"]["WP1227", "DataNode"][
All, #"GraphId" -> #TextLabel &]];
graph = ResourceFunction["WikiPathways"]["WP1227", "Graph", "DeleteAnchors" -> True];](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/3a1756b3d64dae51.png)

![MapThread[
With[
{geneProduct = Function[path,
Normal[
ResourceFunction["WikiPathways"][path, "DataNode"][
Select[MatchQ[#Type, "GeneProduct"] &]][All, #TextLabel &]]
] /@ #1},
BarChart[
Reverse[SortBy[Tally[Flatten[geneProduct]], Last]][[All, 2]],
PlotLabel -> #2, Frame -> True, FrameLabel -> {"GeneProduct", "Frequency"}
]
] &,
{twoPath, Normal[Keys[allPaths[GroupBy["Name"]][TakeLargestBy[Length, 2]]]]}]](https://www.wolframcloud.com/obj/resourcesystem/images/ab8/ab81ab85-9317-4fea-bb15-b96c7609c487/1-0-0/21ddce17e4eb0a6d.png)