Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Retrieve genome track data accessible in the UCSC Genome Browser database
ResourceFunction["UCSCGenomeTrackData"]["Hubs"] gives the dataset of available public hubs to access assembled genomics information. | |
ResourceFunction["UCSCGenomeTrackData"]["hub","genome"] gives the dataset of available genome tracks from a specified hub and genome. | |
ResourceFunction["UCSCGenomeTrackData"]["hub","genome","track"] gives the track dataset for a specified track from a specified hub and genome. | |
ResourceFunction["UCSCGenomeTrackData"]["hub","genome","track","chromosome"] gives the track dataset for a specified track and chromosome from a specified hub and genome. | |
ResourceFunction["UCSCGenomeTrackData"]["hub","genome","track","chromosome",{start,end}] gives the track dataset for a specified track and chromosome from a specified huband genome. Coordinates of the chromosome are given by the start and end positions. | |
ResourceFunction["UCSCGenomeTrackData"]["hub","genome","track",MetaInformation] gives the metainformation on available track properties for a specified track from a specified hub and genome. |
Retrieve the list of public hubs to access genomics information:
| In[1]:= |
| Out[1]= |  |
Retrieve the dataset of available genome tracks from the latest human assemblies in the UCSC Genome Browser database:
| In[2]:= |
| Out[2]= | 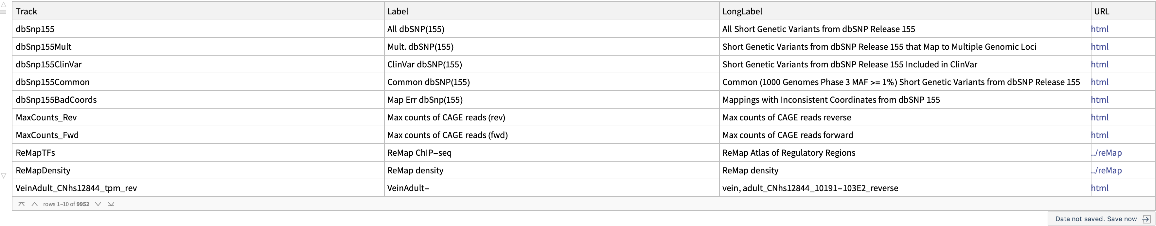 |
Retrieve the known genes in the human X chromosome:
| In[3]:= |
| Out[3]= | 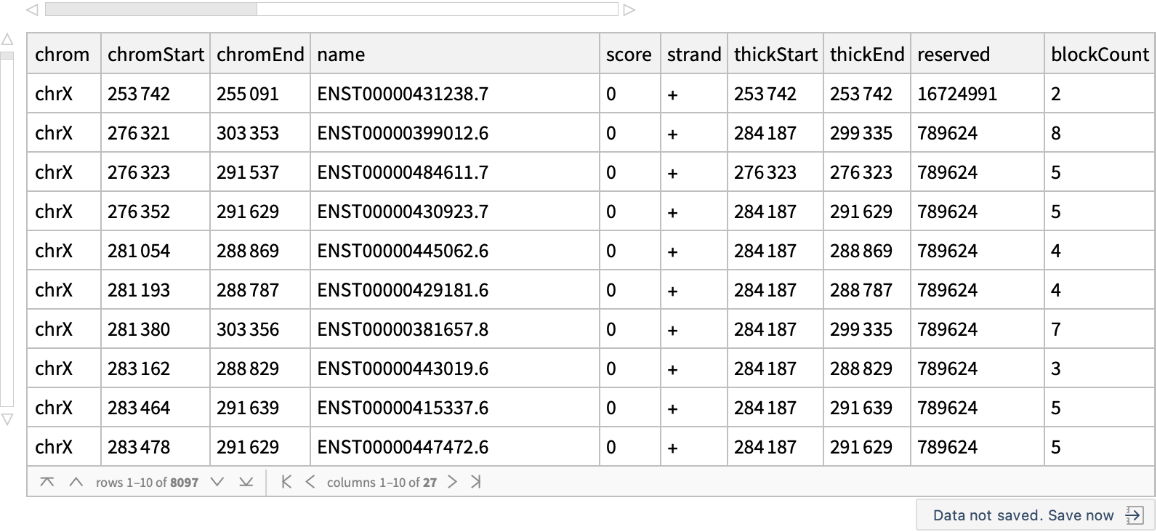 |
Retrieve pathogenicity information for the specified regions of human chromosome 1:
| In[4]:= |
| Out[4]= | 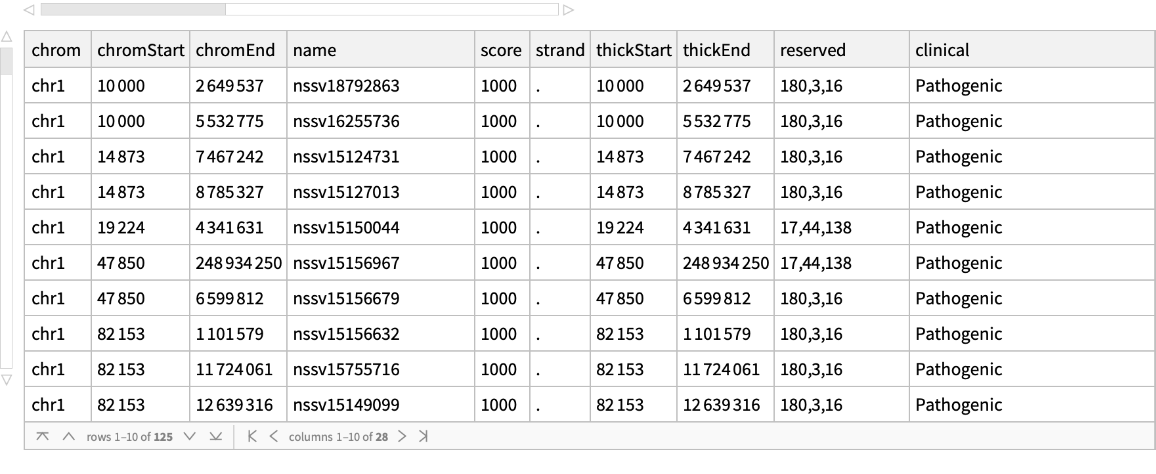 |
Retrieve meta information on properties accessible for a specified genome track:
| In[5]:= | ![ResourceFunction[
"UCSCGenomeTrackData", ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"]["ABC of cellular microRNAome", "hg38", "primary_cells", MetaInformation]](https://www.wolframcloud.com/obj/resourcesystem/images/220/2200f64a-dbc7-4d66-b7bf-b6493db529b5/20c7ce504b73e349.png) |
| Out[5]= | 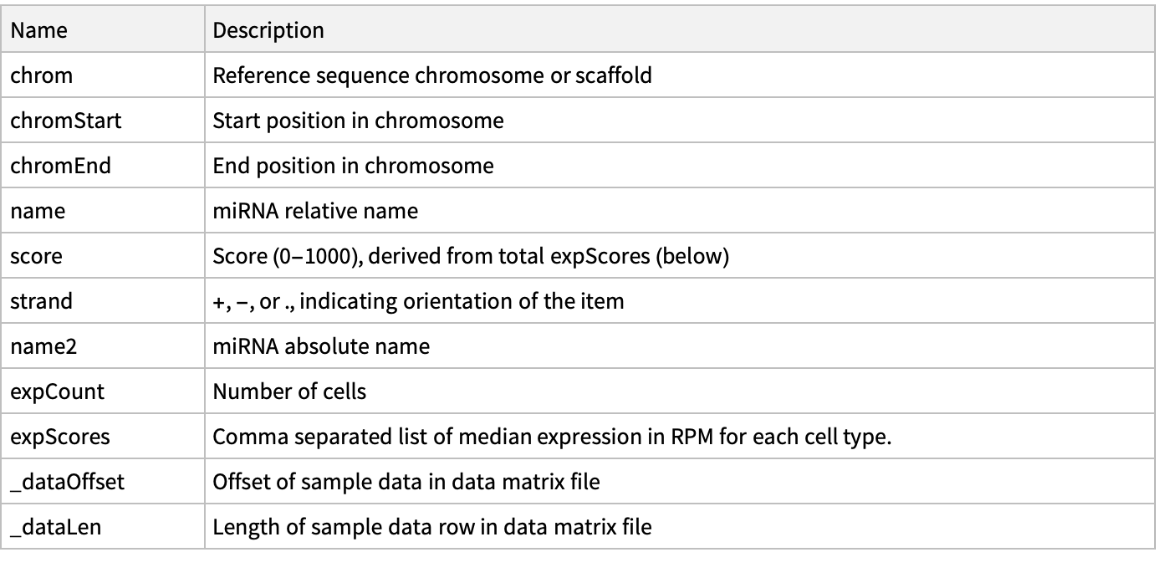 |
Access LIBD Human DLPFC Development hub and retrieve data from the differential expression analysis of the developing human brain:
| In[6]:= |
| Out[6]= | 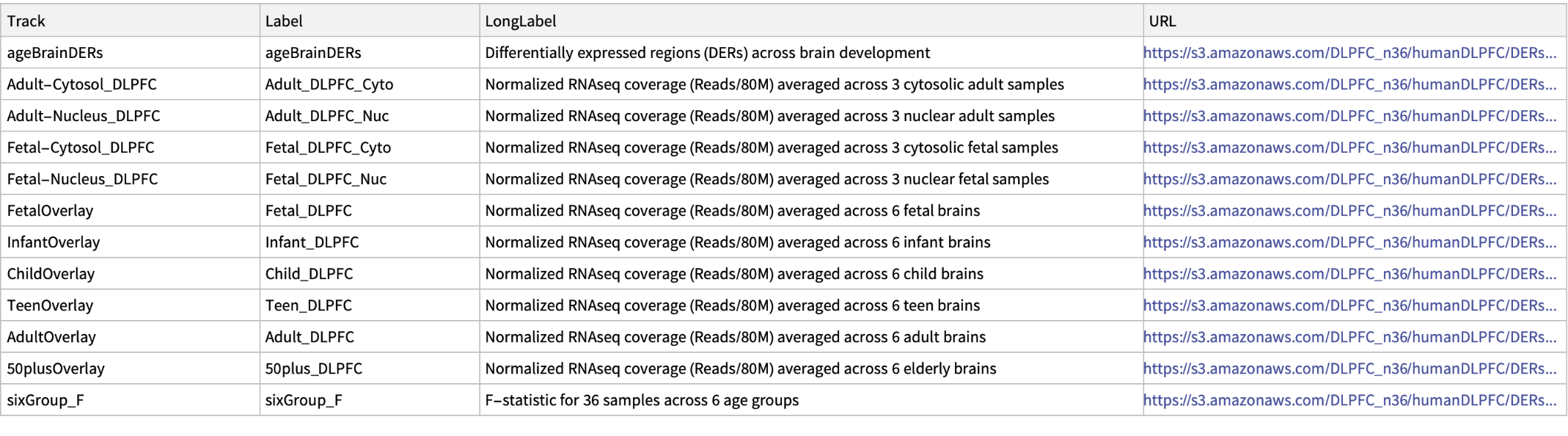 |
Find F-statistics for differential expression regions across six age groups for the chromosome Y:
| In[7]:= |
| Out[7]= | 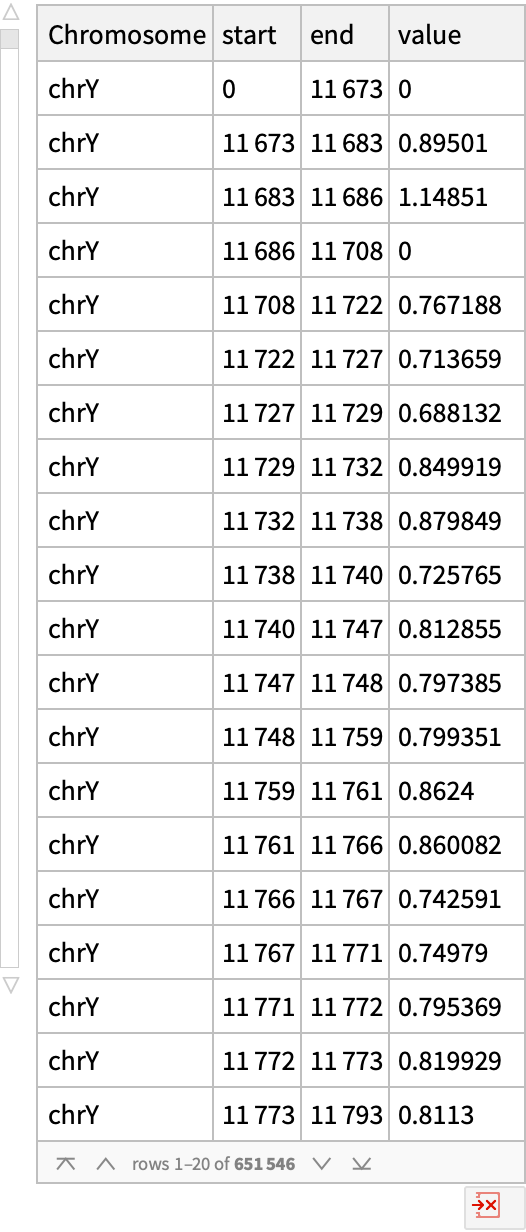 |
Plot the F-values across the length of the chromosome:
| In[8]:= |
| Out[8]= | 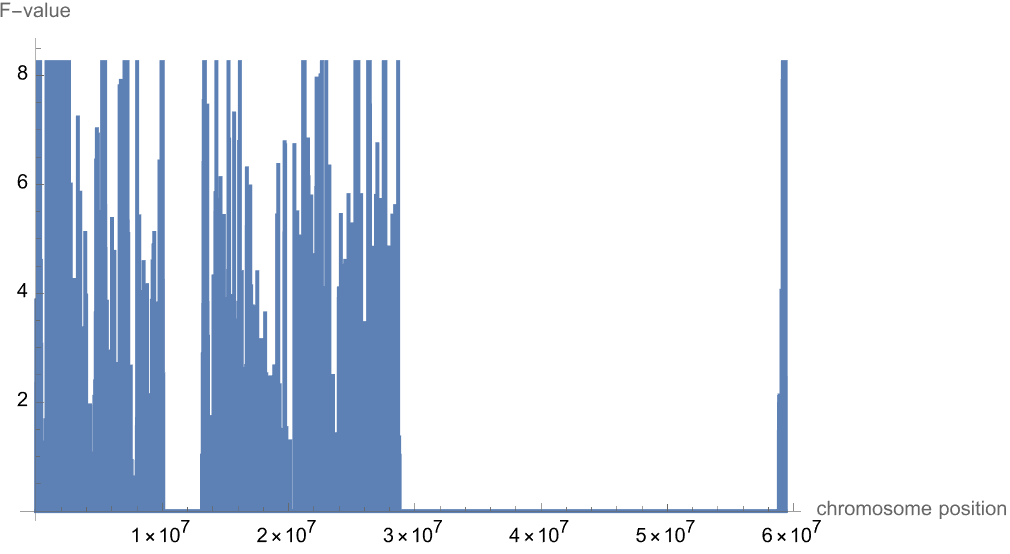 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License