Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Retrieve DNA sequences from the UCSC Genome Browser database
ResourceFunction["UCSCGenomeSequenceData"]["GenomeAssemblies"] gives the dataset of available assembled genomes. | |
ResourceFunction["UCSCGenomeSequenceData"]["genome","chromosome"] gives the DNA sequence from a specified chromosome in the genome. | |
ResourceFunction["UCSCGenomeSequenceData"]["genome","chromosome",{start,end}] gives the DNA sequence from a specified chromosome with the part specified by the start and end positions in the genome. | |
ResourceFunction["UCSCGenomeSequenceData"]["species","chromosome"] gives the DNA sequence from a specified chromosome in the latest available genome of a given species. | |
ResourceFunction["UCSCGenomeSequenceData"]["species","chromosome",{start,end}] gives the DNA sequence from a specified chromosome with the part specified by the start and end positions in the latest available genome of a given species. |
Retrieve available genome assembly information as a dataset:
| In[1]:= |
| Out[1]= | 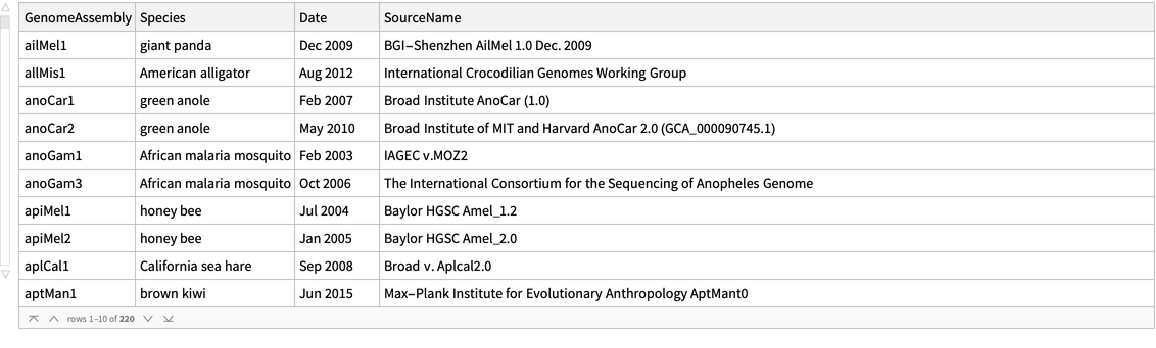 |
Retrieve the DNA sequence from the chromosome in the specified human genome assembly:
| In[2]:= |
| Out[2]= |
Retrieve the DNA sequence from the chromosome in the latest available dog genome assembly:
| In[3]:= |
| Out[3]= |
Repeated sequences are shown in lower-cased letters:
| In[4]:= |
| Out[4]= | 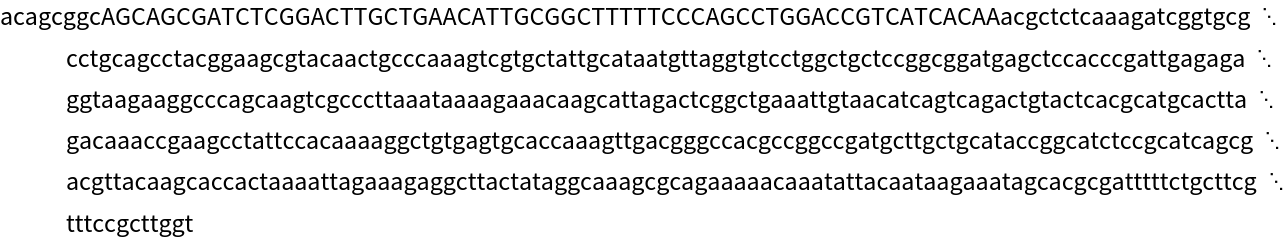 |
Convert the retrieved DNA sequence to the BioSequence object:
| In[5]:= |
| Out[5]= |
Translate a DNA sequence into the corresponding peptide sequences:
| In[6]:= |
| Out[6]= |
Create a molecule and plot it:
| In[7]:= |
| Out[7]= | 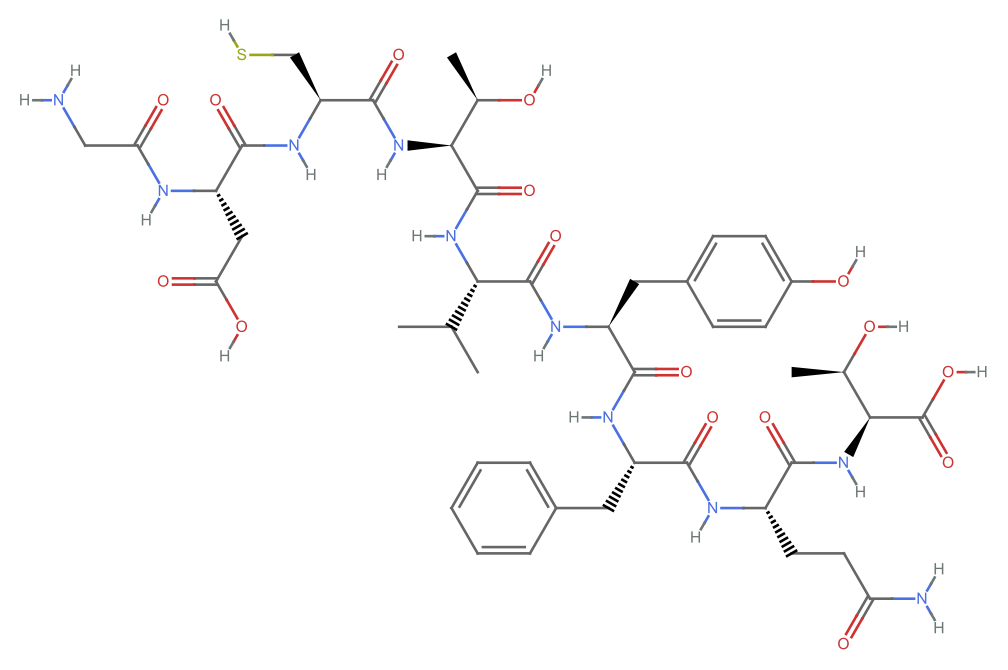 |
Retrieving the entire sequence for a chromosome may take a long time due to the size of data to download:
| In[8]:= |
| Out[8]= |
If the requested end position extends past the end coordinate of the chromosome, the sequence limited to the size of the chromosome is returned:
| In[9]:= |
| Out[9]= |  |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License