Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Find genome information for a given taxonomic species
ResourceFunction["SpeciesGenomeSummary"][species] gives genomic summary information for a specified species entity. | |
ResourceFunction["SpeciesGenomeSummary"][species,property] gives the value of the specified genomic property for the given species. | |
ResourceFunction["SpeciesGenomeSummary"][species,property,format] gives the summary information in a specified format. | |
ResourceFunction["SpeciesGenomeSummary"][species,format] gives all information in the specified format. |
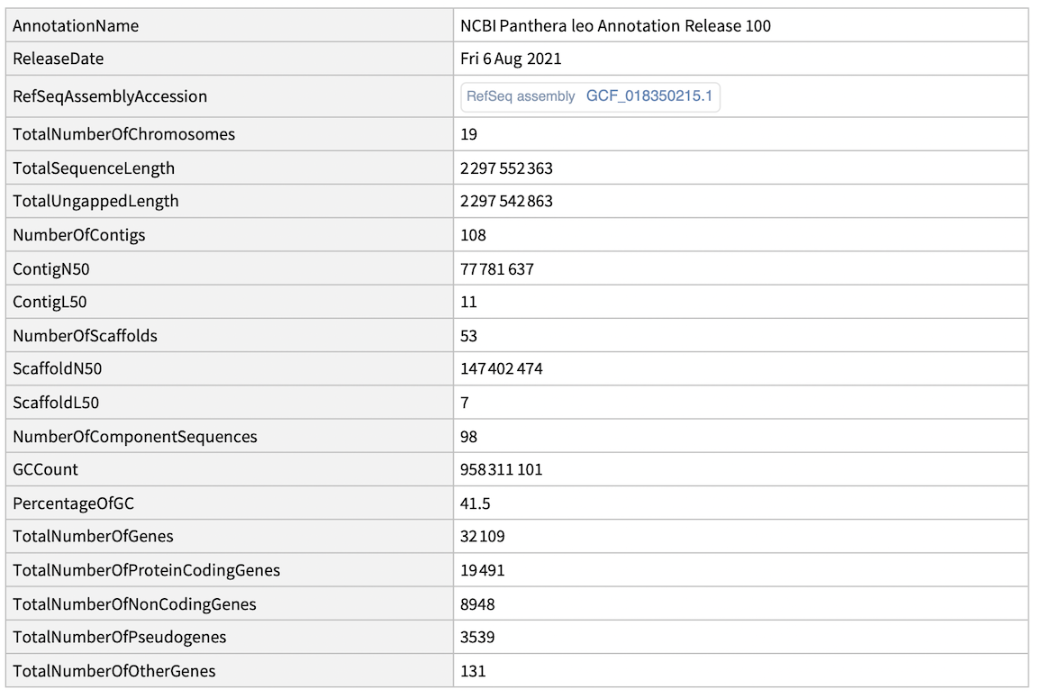
| "AnnotationName" | name of the genome annotation |
| "ReleaseDate" | date of release for the genome asssembly |
| "RefSeqAssemblyAccession" | ExternalIdentifier object representing a RefSeq assebly accession number |
| "TotalNumberOfChromosomes" | total number of chromosomes in the primary assembly |
| "TotalSequenceLength" | total length of sequences including bases and gaps in the primary assembly |
| "TotalUngappedLength" | total length of all top‐level sequences ignoring gaps in the primary assembly; any stretch of 10 or more ambiguous bases (Ns) in a sequence is treated like a gap |
| "NumberOfContigs" | total number of sequence contigs in the primary assembly; any stretch of 10 or more ambiguous bases (Ns) in a sequence is treated as a gap between two contigs in a scaffold when counting contigs and calculating contig N50 & L50 values |
| "ContigN50" | length such that sequence contigs of this length or longer include half the bases of the primary assembly |
| "ContigL50" | number of sequence contigs that are longer than, or equal to, the N50 length and therefore include half the bases of the primary assembly |
| "NumberOfScaffolds" | number of scaffolds including placed, unlocalized, unplaced, alternate loci and patch scaffolds in the primary assembly |
| "ScaffoldN50" | length such that scaffolds of this length or longer include half the bases of the primary assembly |
| "ScaffoldL50" | number of scaffolds that are longer than, or equal to, the N50 length and therefore include half the bases of the primary assembly |
| "NumberOfComponentSequences" | total number of component Whole Genome Shotgun (WGS) or clone sequences in the primary assembly |
| "GCCount" | number of guanine (G) or cytosine (C) bases in the primary assembly |
| "PercentageOfGC" | percentage of guanine (G) or cytosine (C) bases in the primary assembly |
| "TotalNumberOfGenes" | total number of reported genes in the primary assembly |
| "TotalNumberOfProteinCodingGenes" | total number of protein coding genes in the primary assembly |
| "TotalNumberOfNonCodingGenes" | total number of non‐coding genes in the primary assembly |
| "TotalNumberOfPseudogenes" | total number of pseudogene in the primary assembly |
| "TotalNumberOfOtherGenes" | total number of genes other than protein coding, non‐coding, and pseudo‐ genes in the primary assembly |
| "Association" | Association of species entities and entity-property values |
| "Dataset" | Dataset in which the specified species entities are keys, and values are an Association of property names and entity-property values |
Get the genome report for lions:
| In[1]:= |
Explore a specific genomic property:
| In[2]:= |
| Out[2]= |
Get gene information as an Association:
| In[3]:= | ![ResourceFunction["SpeciesGenomeSummary"][
Entity["TaxonomicSpecies", "GorillaGorilla::57mg3"], {"TotalNumberOfGenes", "TotalNumberOfProteinCodingGenes", "TotalNumberOfNonCodingGenes"}, "Association"]](https://www.wolframcloud.com/obj/resourcesystem/images/5d1/5d17cfcb-088b-4cca-8373-48f99450915e/736828b4bf917454.png) |
| Out[3]= |
Compare genomic characteristics for common fruit plants:
| In[4]:= | ![fruit = {Entity["TaxonomicSpecies", "PrunusPersica::76q6r"], Entity["TaxonomicSpecies", "MalusDomestica::k7rps"], Entity["TaxonomicSpecies", "AnanasComosus::7524s"], Entity["TaxonomicSpecies", "CitrusSinensis::fdd23"], Entity["TaxonomicSpecies", "VitisVinifera::v8884"], Entity["TaxonomicSpecies", "MusaAcuminata::35k78"]};](https://www.wolframcloud.com/obj/resourcesystem/images/5d1/5d17cfcb-088b-4cca-8373-48f99450915e/02400f5723ec45ba.png) |
| In[5]:= |
| Out[5]= | 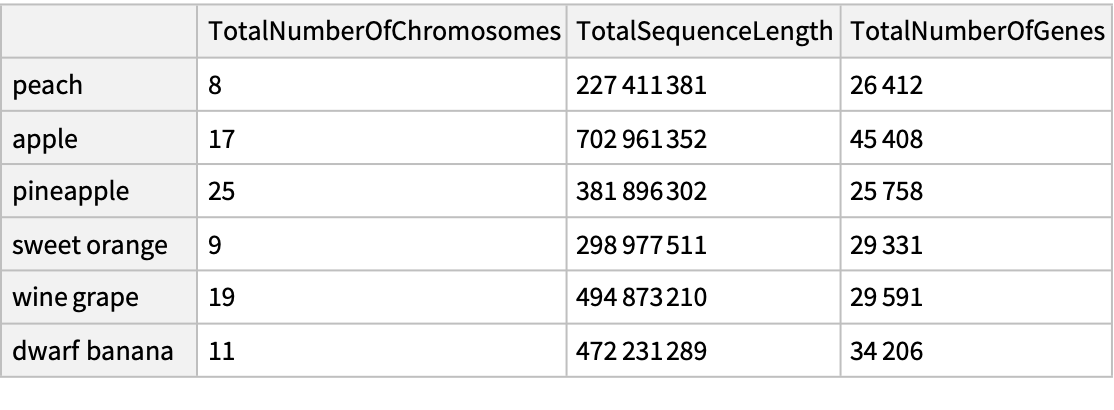 |
Plot the total genome length against the number of chromosomes:
| In[6]:= |
| Out[6]= | 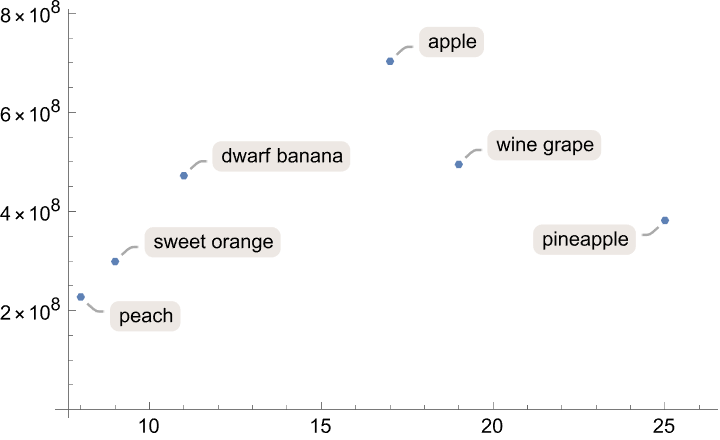 |
Genome information is available for selected species only. Trying to visualize a higher rank taxon returns Missing:
| In[7]:= |
| Out[7]= |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License