Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Interactively manipulate the conformation of a flexible molecule and paste results along the way
ResourceFunction["RotateBonds3D"][mol] generates an interactive GUI that allows the user to dynamically rotate bonds in the molecule mol. |
| "AnchorAtom" | Automatic | index of atom at which the spanning tree of rotatable bonds is anchored (rooted), or {anchor,azimuth} where azimuth is the index of an atom adjacent to anchor}. |
| "DiagramSize" | Automatic | the effective ImageSize for the schematic diagram showing the rotatable bonds and atom indices |
| IncludeHydrogens | False | controls the display of hydrogen atoms and the inclusion of rotatable bonds terminating in, for example, a methyl group |
Rotate butane about its central C-C bond and paste its gauche and anti conformations:
| In[1]:= |
| Out[1]= | 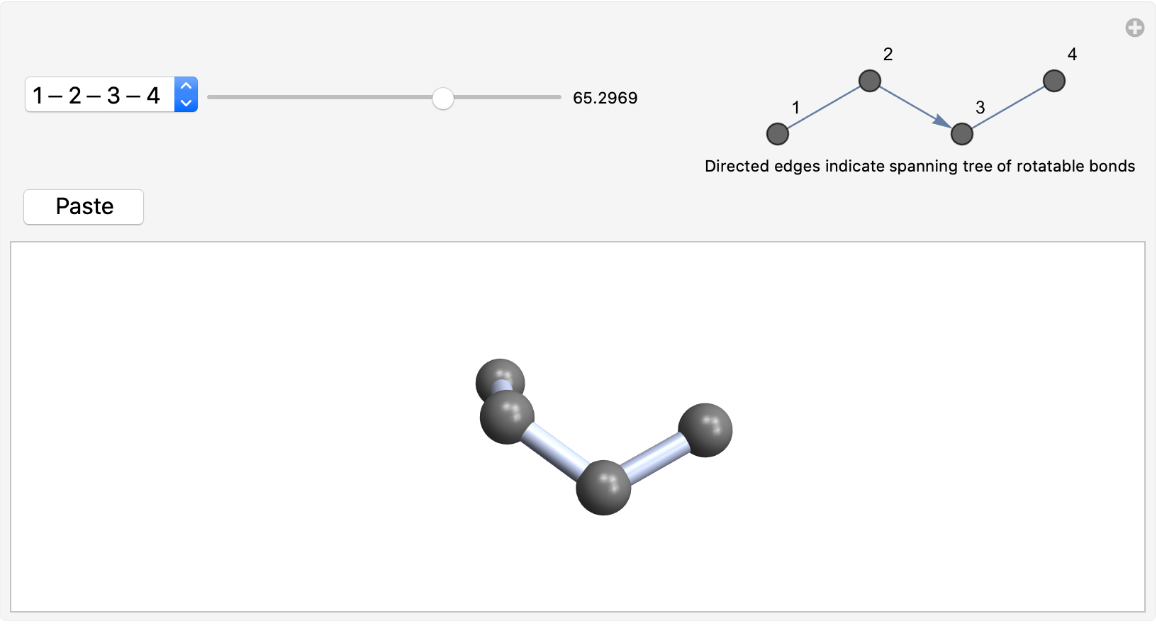 |
Copy each pasted Molecule and visualize the results:
| In[2]:= | ![Molecule[{"C", "C", "C", "C", "H", "H", "H", "H", "H", "H", "H", "H", "H", "H"}, {
Bond[{1, 2}, "Single"],
Bond[{2, 3}, "Single"],
Bond[{3, 4}, "Single"],
Bond[{1, 5}, "Single"],
Bond[{1, 6}, "Single"],
Bond[{1, 7}, "Single"],
Bond[{2, 8}, "Single"],
Bond[{2, 9}, "Single"],
Bond[{3, 10}, "Single"],
Bond[{3, 11}, "Single"],
Bond[{4, 12}, "Single"],
Bond[{4, 13}, "Single"],
Bond[{4, 14}, "Single"]}, {AtomCoordinates -> QuantityArray[
StructuredArray`StructuredData[{14, 3}, {CompressedData["
1:eJwBYQGe/iFib1JlAgAAAA4AAAADAAAAO7tDAWLr+L8edherbDDiPw5mZrmh
5sC/AAMXR8Bb5r8pOajSXmfjv2zF0lzD2tM/tvfBG8Bb5j9sqerVXmfjv1N/
bk3D2tO/lm389MUx+D9r+r4YkkDkP0RHTIfasp4/j0xZ4ZoX+r8q1qoispzj
P/DgKiFikvO/rCtNXGePBMCee3QCO9bdPwH7PQbPX9E/5v8KFBxw8r/05FB/
o034P1hrgfrGE80/sXRlqEhL8799Hxok7JL4v+AcDzsW5Zk/DiJhX4rj479m
cz4vHITjv1qBce6ydfY/xWENkj/z4z+VlBXnf4/mv2VLuTMCZPa/nPgYk5rW
8z/urmRMXdL3v+BIIAVoRqo/NFKGGeJP+T+C8CttA7ToP2pfVTaQ0PE/cvOL
NCMo8T/kJL2/yIr4P3KnFdiLFNq/b6VBeEo6BEAomdlurR7hP15QghVbXde/
gqmuyQ==
"], "Angstroms", {{1}, {2}}}]]}] // MoleculePlot3D[#, ImageSize -> Small, PlotLabel -> Style["gauche", Italic]] &](https://www.wolframcloud.com/obj/resourcesystem/images/2b4/2b412c96-2f1c-453e-84f6-736eea35f1ab/4ca36145be23c39e.png) |
| Out[2]= | 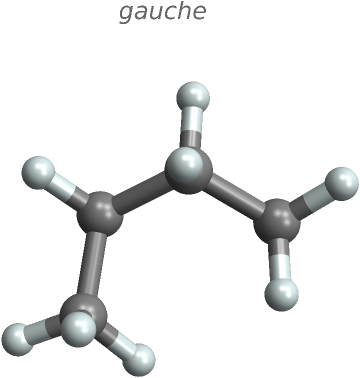 |
| In[3]:= | ![Molecule[{"C", "C", "C", "C", "H", "H", "H", "H", "H", "H", "H", "H", "H", "H"}, {
Bond[{1, 2}, "Single"],
Bond[{2, 3}, "Single"],
Bond[{3, 4}, "Single"],
Bond[{1, 5}, "Single"],
Bond[{1, 6}, "Single"],
Bond[{1, 7}, "Single"],
Bond[{2, 8}, "Single"],
Bond[{2, 9}, "Single"],
Bond[{3, 10}, "Single"],
Bond[{3, 11}, "Single"],
Bond[{4, 12}, "Single"],
Bond[{4, 13}, "Single"],
Bond[{4, 14}, "Single"]}, {AtomCoordinates -> QuantityArray[
StructuredArray`StructuredData[{14, 3}, {CompressedData["
1:eJwBYQGe/iFib1JlAgAAAA4AAAADAAAAO7tDAWLr+L8edherbDDiPw5mZrmh
5sC/AAMXR8Bb5r8pOajSXmfjv2zF0lzD2tM/tvfBG8Bb5j9sqerVXmfjv1N/
bk3D2tO/PPf4EWLr+D+pqvcylX/8vw5rG4Oh5sA/j0xZ4ZoX+r8q1qoispzj
P/DgKiFikvO/rCtNXGePBMCee3QCO9bdPwH7PQbPX9E/5v8KFBxw8r/05FB/
o034P1hrgfrGE80/sXRlqEhL8799Hxok7JL4v+AcDzsW5Zk/DiJhX4rj479m
cz4vHITjv1qBce6ydfY/KvMsGL5p8z+yNvAueDfVP8APDE0MJrK/NJi/R60J
4z8uIjbuDofkvyJxFhyja/a/tIq1rynm8D8I772FxdwFwHhN2adsIbK/3fYJ
iKqU/D+KPYHRVbH7vzs1+cSLNfM/UGS9JJ0QBEDvf7a6TG38v8D2e11ITNq/
KYmwsw==
"], "Angstroms", {{1}, {2}}}]]}] // MoleculePlot3D[#, ImageSize -> Small, PlotLabel -> Style["anti", Italic]] &](https://www.wolframcloud.com/obj/resourcesystem/images/2b4/2b412c96-2f1c-453e-84f6-736eea35f1ab/1b2b25e167ae7a96.png) |
| Out[3]= | 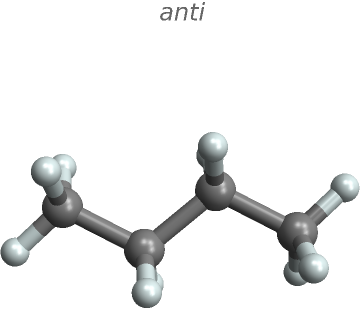 |
Bonds in rings are excluded from consideration:
| In[4]:= |
| Out[4]= | 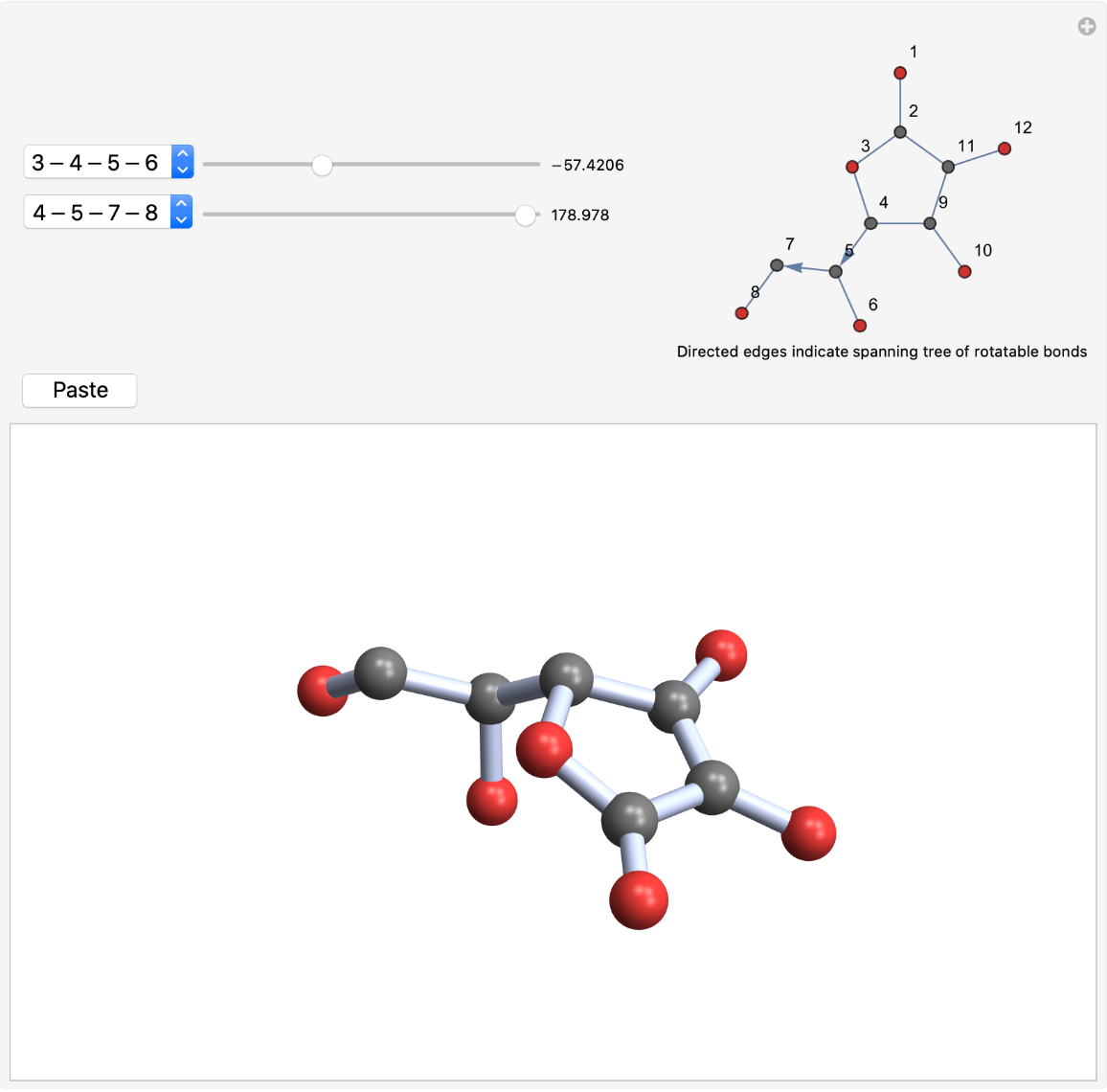 |
By default, the spanning tree of rotatable bonds is anchored at the atom with the highest ClosenessCentrality, which tends to be in the middle of the molecule:
| In[5]:= |
| Out[5]= | 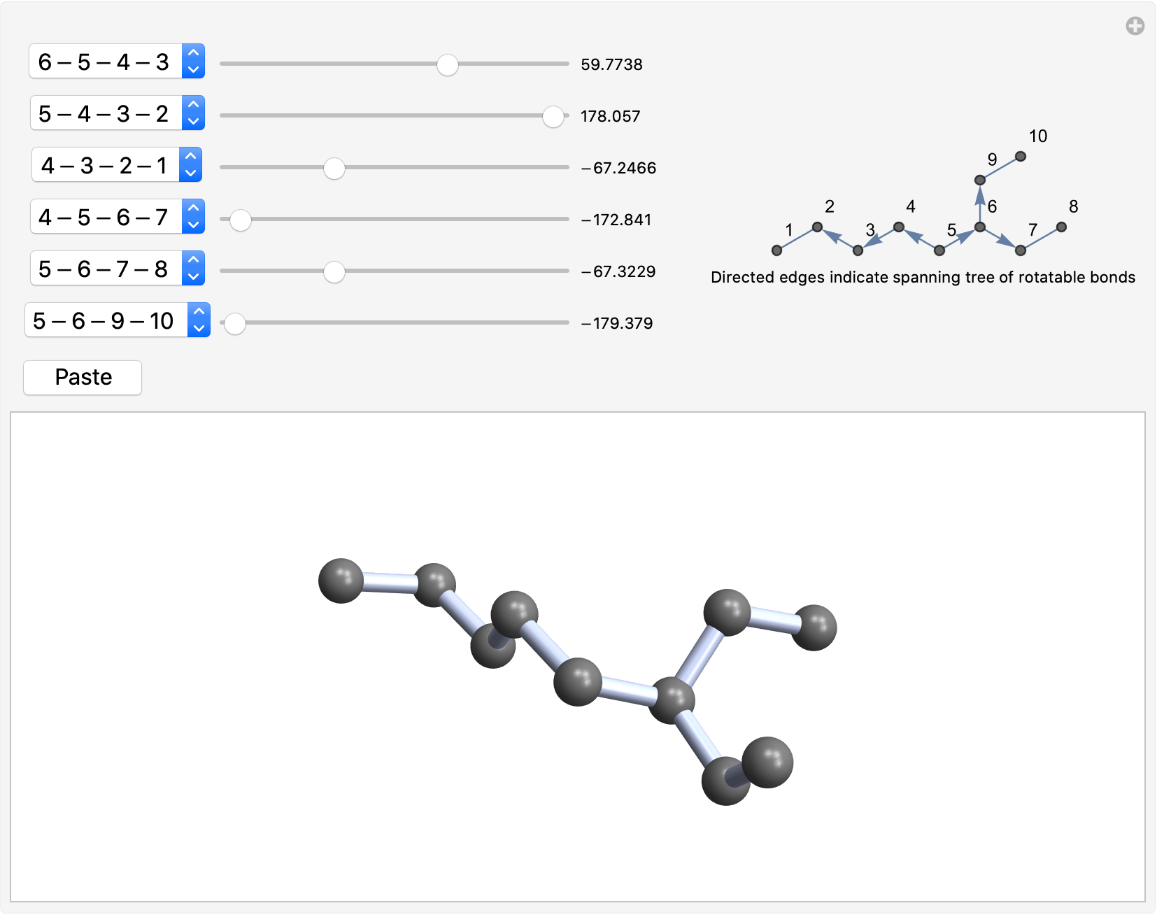 |
Anchor the spanning tree at one end:
| In[6]:= |
| Out[6]= | 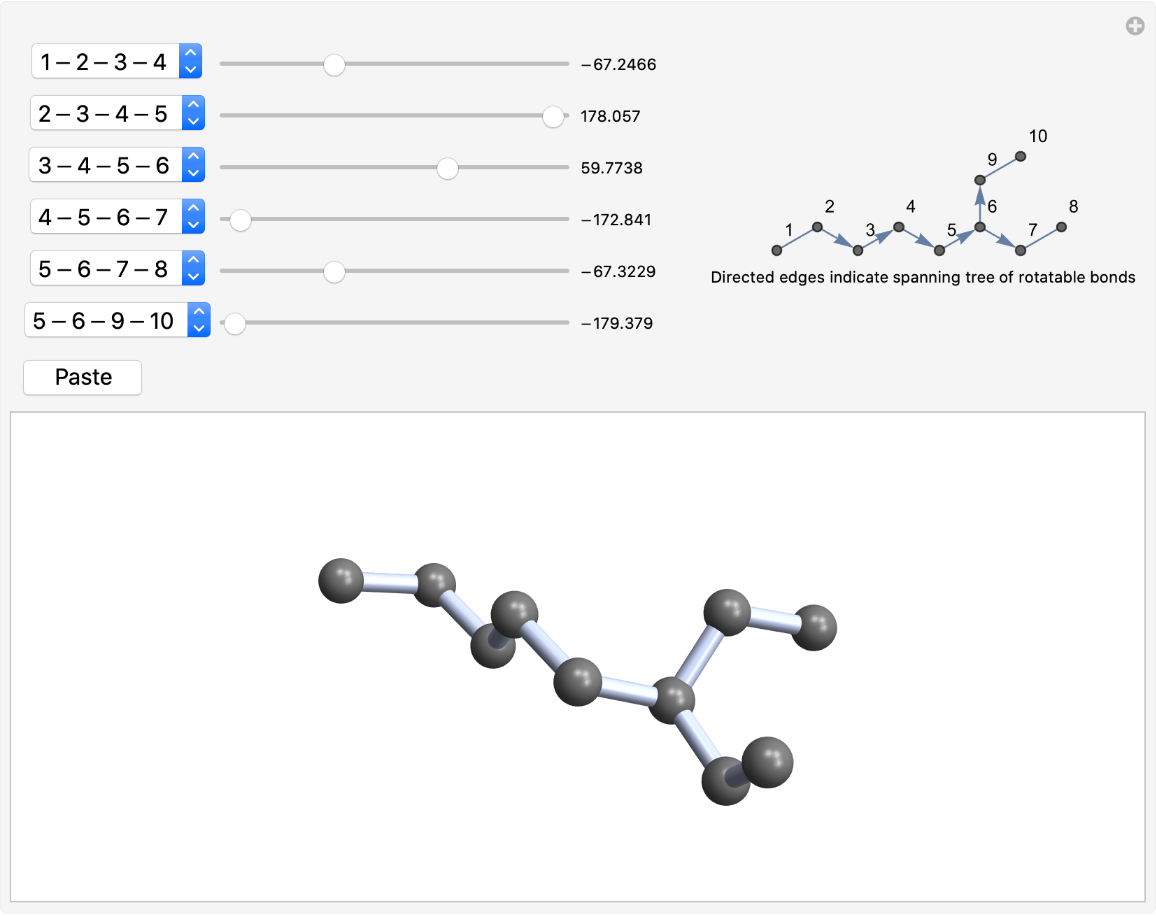 |
When the anchor atom was chosen automatically, the initial direction of the spanning was toward atom 4. Use the two-atom form to change the direction:
| In[7]:= |
| Out[7]= | 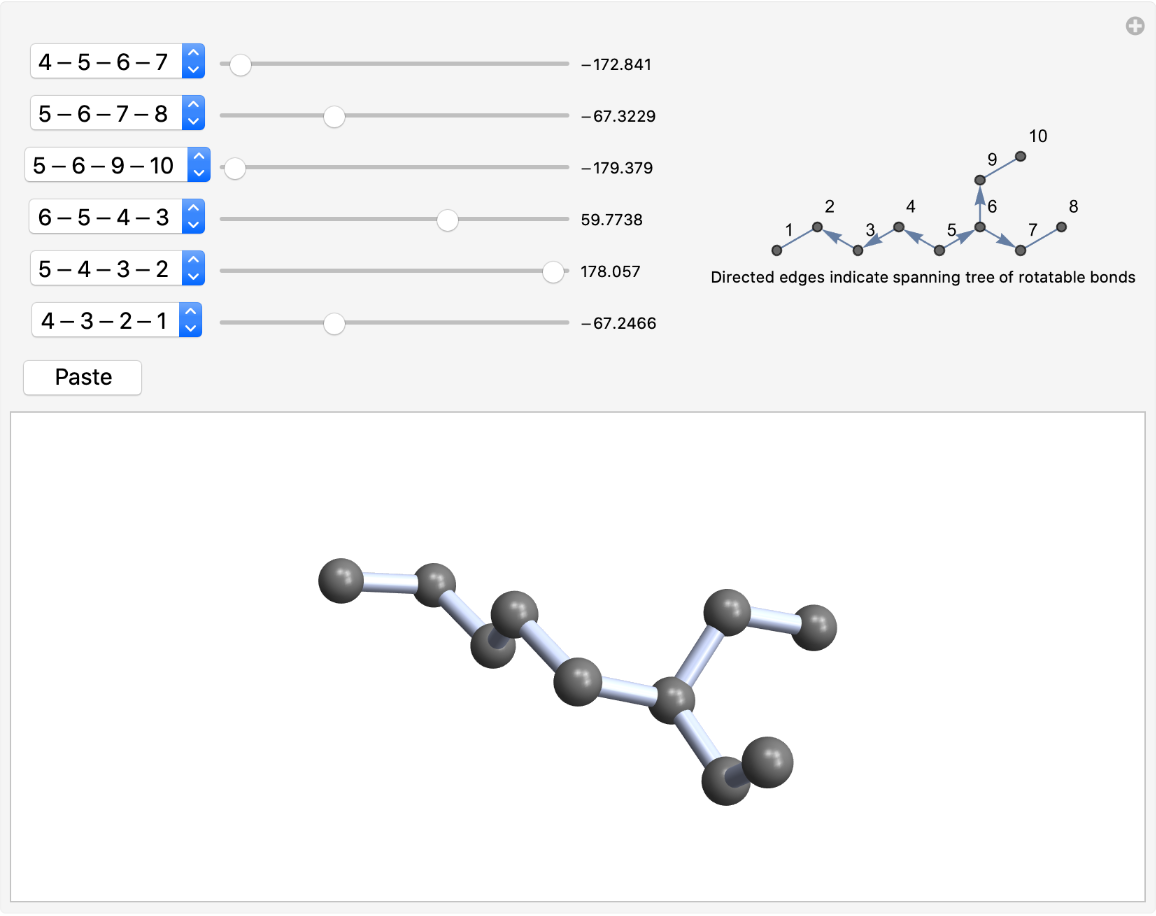 |
Specify the size for the schematic diagram:
| In[8]:= |
| Out[8]= | 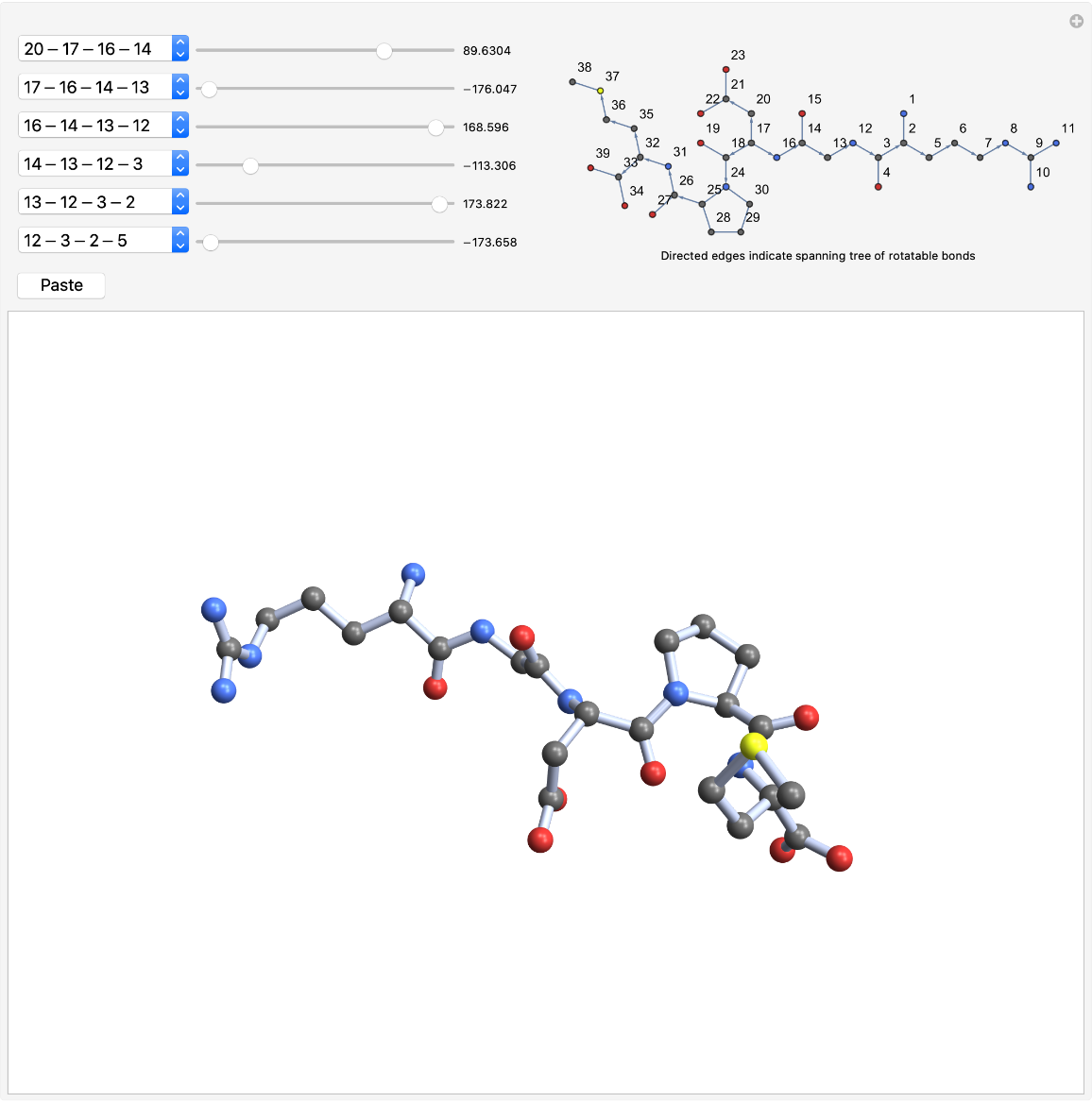 |
By default, hydrogen atoms are not shown:
| In[9]:= |
| Out[9]= | 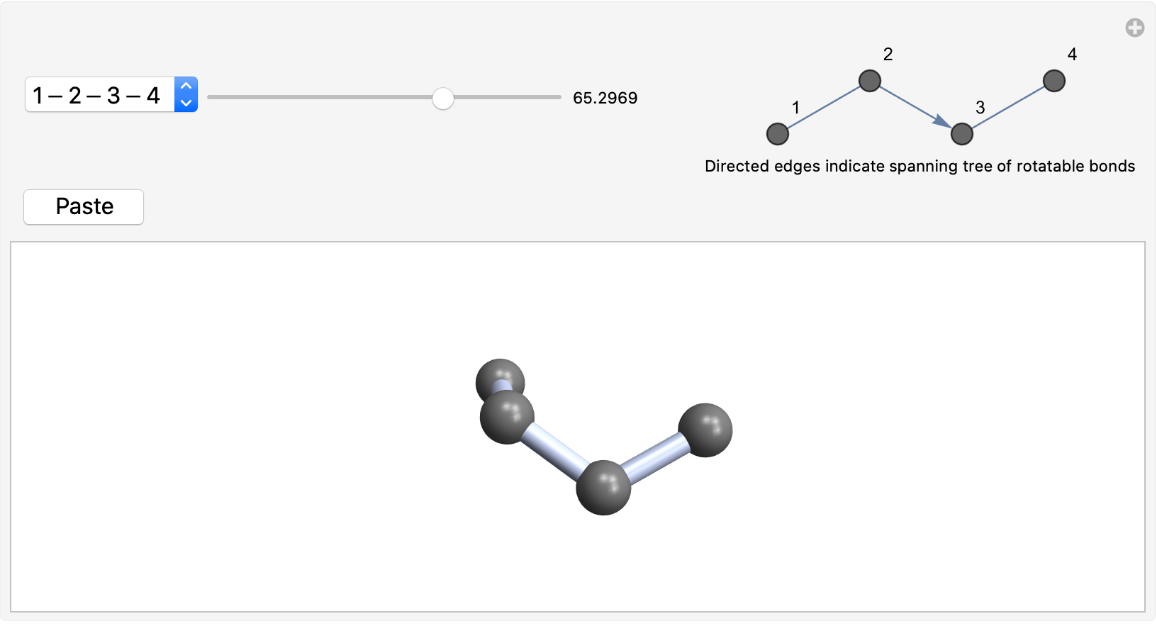 |
Displaying the hydrogen atoms also permits more bonds to be rotatable:
| In[10]:= |
| Out[10]= | 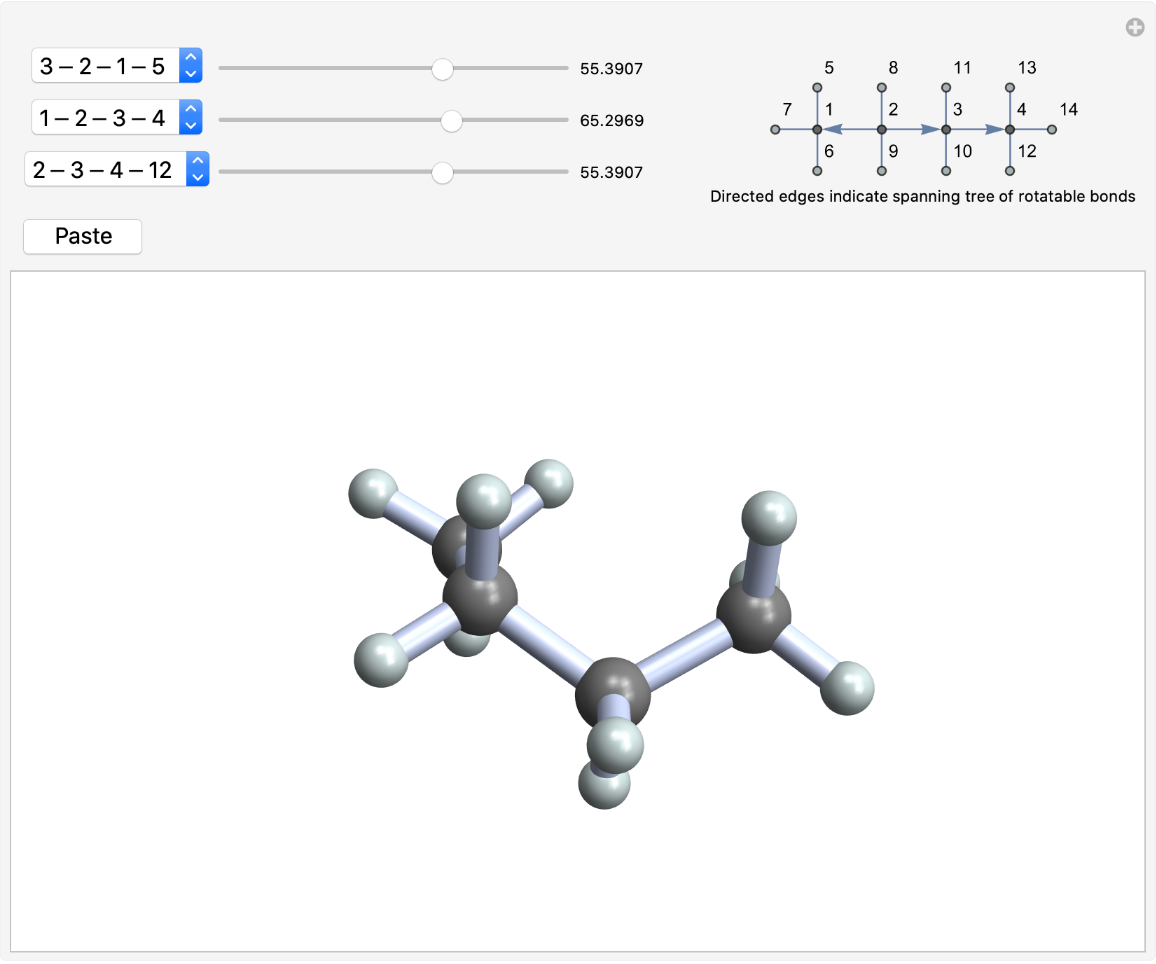 |
Cyclic molecules like cyclohexane have no rotatable bonds, and thus no sliders are presented in RotateBonds3D:
| In[11]:= |
| Out[11]= |  |
RotateBonds3D is a good way to make peptide turns. Start with a tetrapeptide:
| In[12]:= |
| Out[12]= | 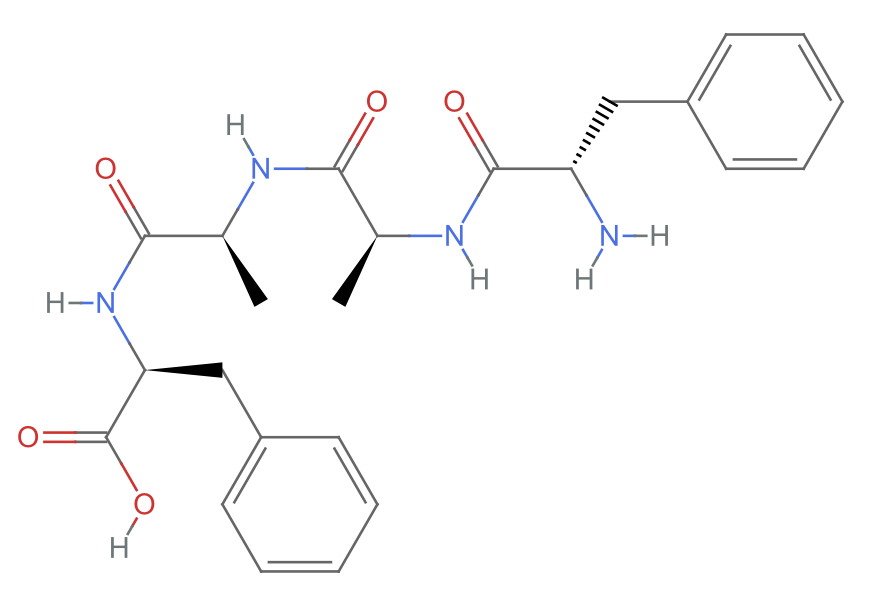 |
Give the structure a type II β-turn. Set the torsion angles according to the values in the table (starting from the bottom of the menus):
| In[13]:= | ![Grid[{{"peptide angle", "torsion angle", "value"},
{"\!\(\*SubscriptBox[\(\[Psi]\), \(i\)]\)", "5-2-3-12", -60},
{"\!\(\*SubscriptBox[\(\[Phi]\), \(i + 1\)]\)", "2-3-12-13", -60},
{"\!\(\*SubscriptBox[\(\[Psi]\), \(i + 1\)]\)", "12-13-14-17", 120},
{"\!\(\*SubscriptBox[\(\[Phi]\), \(i + 2\)]\)", "14-17-18-19", 80},
{"\!\(\*SubscriptBox[\(\[Psi]\), \(i + 2\)]\)", "17-18-19-22", 0},
{"\!\(\*SubscriptBox[\(\[Phi]\), \(i + 3\)]\)", "19-22-23-24", 60}
}, Dividers -> {None, {False, True, False}}, Spacings -> {2, Automatic}]](https://www.wolframcloud.com/obj/resourcesystem/images/2b4/2b412c96-2f1c-453e-84f6-736eea35f1ab/1e442be39214566c.png) |
| Out[13]= | 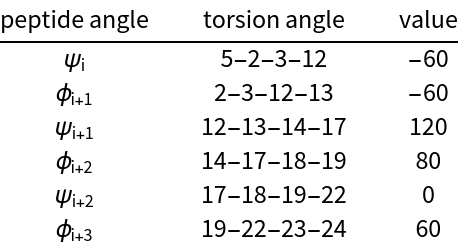 |
| In[14]:= |
| Out[14]= | 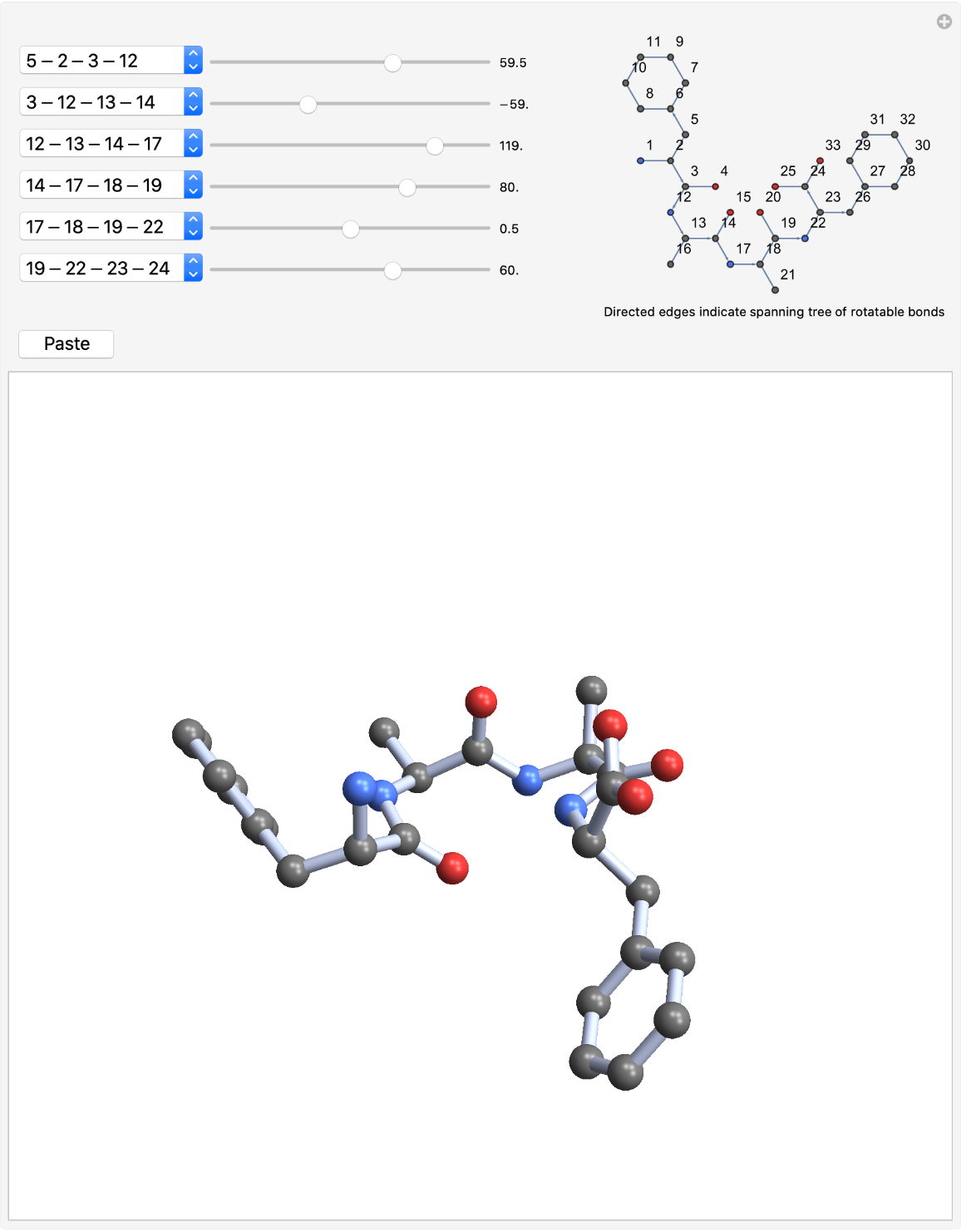 |
| In[15]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/2533d0f9-c151-4744-b370-d9853fe868a0"]](https://www.wolframcloud.com/obj/resourcesystem/images/2b4/2b412c96-2f1c-453e-84f6-736eea35f1ab/13dc1cdcd2ea2349.png) |
| Out[15]= | 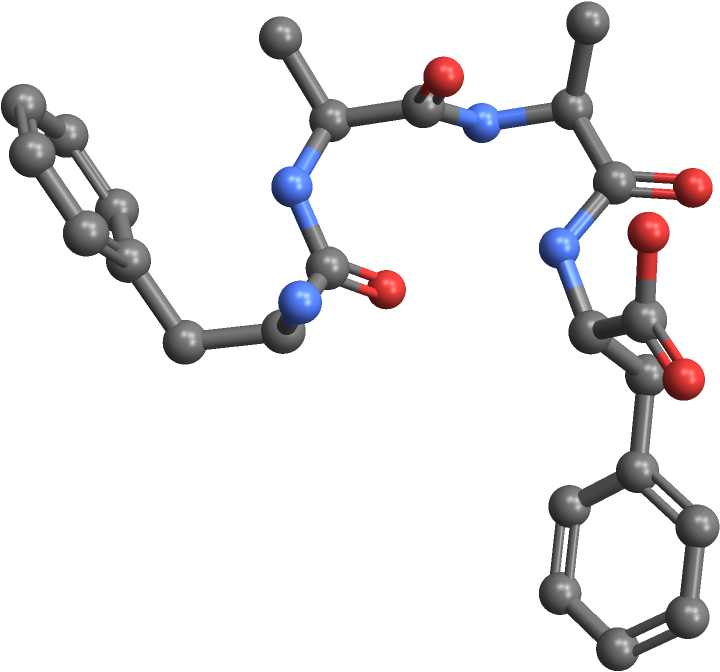 |
This work is licensed under a Creative Commons Attribution 4.0 International License