Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Calculate the network-based inference (NBI) connection score between two vertices in a bipartite graph
ResourceFunction["NetworkBasedInferenceScore"][g,d,t] gives the network based inference (NBI) score between the drug vertex d and the target vertex t in the graph g. |
Create a simple network of pharmaceutical drugs and their viral targets:
| In[1]:= |
| Out[1]= | 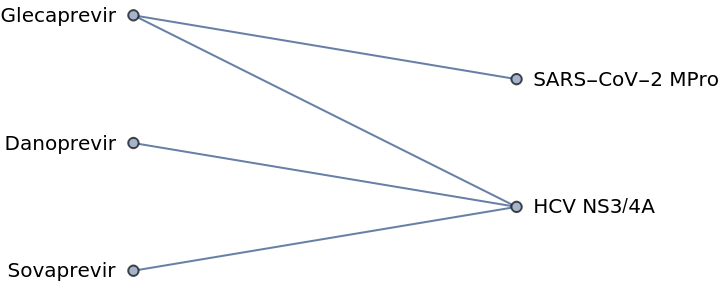 |
Calculate the NBI score between a drug and target vertex:
| In[2]:= |
| Out[2]= |
A bipartite graph:
| In[3]:= |
| Out[3]= | 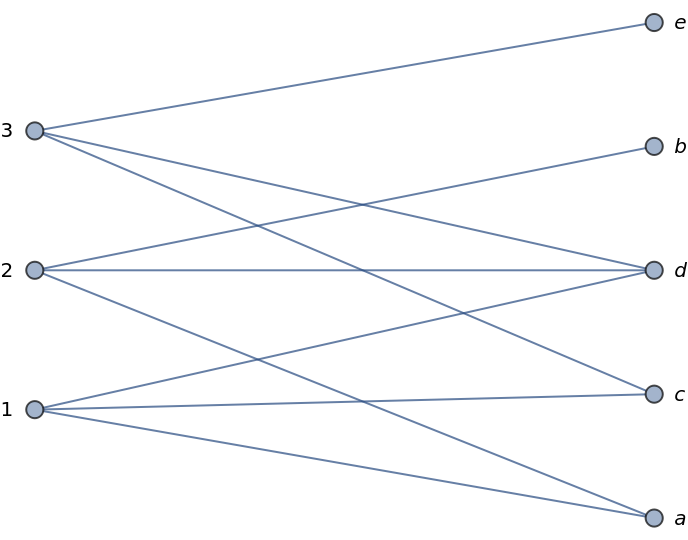 |
The NBI score can be calculated for pairs of vertices that have an edge between them as well:
| In[4]:= |
| Out[4]= |
Network edges can be labeled with the NBI scores:
| In[5]:= |
| In[6]:= | ![g3 = Graph[{"Sovaprevir", "Danoprevir", "HCV NS3/4A", "SARS-CoV-2 MPro"}, {
UndirectedEdge["Sovaprevir", "HCV NS3/4A"],
UndirectedEdge["Danoprevir", "HCV NS3/4A"],
UndirectedEdge["Glecaprevir", "HCV NS3/4A"],
UndirectedEdge["Glecaprevir", "SARS-CoV-2 MPro"],
UndirectedEdge["Sovaprevir", "SARS-CoV-2 MPro"],
UndirectedEdge["Danoprevir", "SARS-CoV-2 MPro"]}, GraphLayout -> "BipartiteEmbedding", VertexLabels -> All, EdgeLabels -> Table[i -> Placed[N[
ResourceFunction["NetworkBasedInferenceScore"][g1, i[[1]], i[[2]]]], 1/4], {i, Join[EdgeList[g1], noEdge]}], EdgeStyle -> Table[i -> Dashed, {i, noEdge}]]](https://www.wolframcloud.com/obj/resourcesystem/images/886/88607ecd-4988-47e9-b21c-1141711cd72c/0da4349694fc4230.png) |
| Out[6]= | 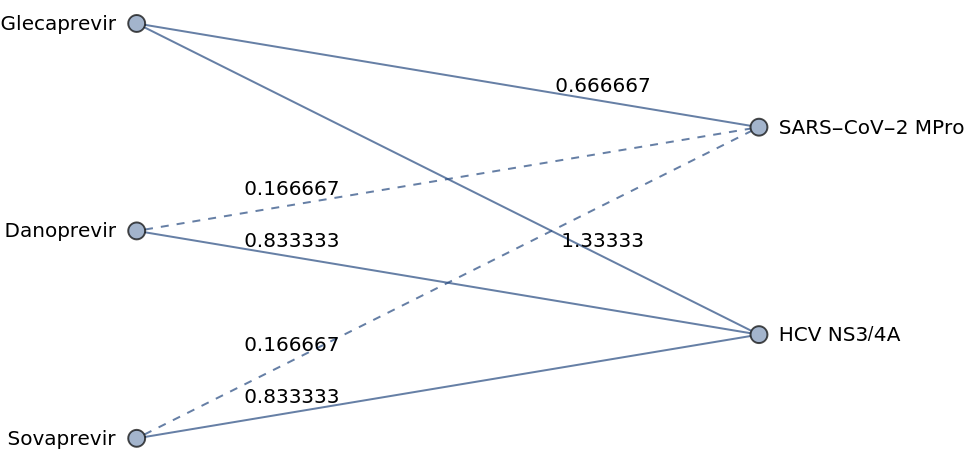 |
This work is licensed under a Creative Commons Attribution 4.0 International License