Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Simulate the evolution of a Petri net configuration as a multiway system
ResourceFunction["MultiwayPetriNet"][pnet,n] generates the results of n steps in the evolution of the multiway Petri net configuration defined by the PetriNetObject expression pnet. | |
ResourceFunction["MultiwayPetriNet"][p,t,a,init,n] generates the results of n steps in the evolution of the multiway Petri net defined by the list of places p, the list of transitions t and the list of arcs a, with initial token distribution init. | |
ResourceFunction["MultiwayPetriNet"][assoc,init,n] generates the results of n steps in the evolution of the multiway Petri net defined by the association of places, transitions and arcs assoc, with initial token distribution init. | |
ResourceFunction["MultiwayPetriNet"][…,"prop"] gives the property "prop" for the specified multiway Petri net evolution. | |
ResourceFunction["MultiwayPetriNet"][pnet→sel,n,…] uses the function sel to select which of the events obtained at each step to include in the evolution. |
| "Sequential" | applies the first possible replacement (sequential substitution system) |
| "Random" | applies a random replacement |
| {"Random",n} | applies n randomly chosen replacements |
| "AllStatesList" | the list of all states generated at each successive step |
| "StatesCountsList" | the number of distinct states generated at each successive step |
| "AllStatesListUnmerged" | the list of all states without any merging |
| "PredecessorRulesList" | the list of states and their corresponding predecessor states at each successive step |
| "EvolutionGraph" | graph formed by the evolution process, with no merging between different time steps |
| "EvolutionGraphStructure" | evolution graph without labeling |
| "EvolutionGraphFull" | graph formed by the evolution process, including equivalent events |
| "EvolutionGraphFullStructure" | full evolution graph without labeling |
| "EvolutionGraphUnmerged" | graph formed by the evolution process, with no merging of equivalent states |
| "EvolutionGraphUnmergedStructure" | unmerged evolution graph without labeling |
| "EvolutionGraphWeighted" | graph formed by the evolution process, with edges weighted by event multiplicity |
| "EvolutionGraphWeightedStructure" | weighted evolution graph without labeling |
| "StatesGraph" | graph of how each distinct state leads to other states |
| "StatesGraphStructure" | states graph without labeling |
| "AllEventsList" | the list of all events that occur at each successive step |
| "EvolutionEventsGraph" | graph showing the evolution process with updating events explicitly included |
| "EvolutionEventsGraphStructure" | evolution events graph without labeling |
| "CausalGraph" | graph of all causal relations between updating events |
| "CausalGraphStructure" | causal graph without labeling |
| "EvolutionCausalGraph" | combined graph of evolution process and causal relationships between events |
| "EvolutionCausalGraphStructure" | evolution causal graph without labeling |
| "CausalGraphInstances" | list of distinct causal graphs for all possible choices of event sequences |
| "CausalGraphStructureInstances" | causal graph instances without labeling |
| "EvolutionCausalGraphInstances" | list of distinct evolution causal graphs for all possible choices of event sequences |
| "EvolutionCausalGraphStructureInstances" | evolution causal graph instances without labeling |
| "BranchPairsList" | list of all branch pairs (i.e. critical pairs) generated in the states graph |
| "NewBranchPairsList" | list of all new branch pairs generated at each successive step |
| "EvolutionBranchPairsList" | list of all branch pairs generated in the evolution graph |
| "NewEvolutionBranchPairsList" | list of all new evolution branch pairs generated at each successive step |
| "BranchPairEventsList" | list of all events yielding branch pairs |
| "NewBranchPairEventsList" | list of all events yielding new branch pairs at each successive step |
| "EvolutionBranchPairEventsList" | list of all events yielding evolution branch pairs |
| "NewEvolutionBranchPairEventsList" | list of all events yielding new evolution branch pairs at each successive step |
| "BranchialGraph" | graph of branch pair ancestry at a given step |
| "BranchialGraphStructure" | branchial graph without labeling |
| "AllStatesBranchialGraph" | graph of branch pair ancestry across all steps |
| "AllStatesBranchialGraphStructure" | all states branchial graph without labeling |
| "EventBranchialGraph" | graph of branch pair event ancestry at a given step |
| "EventBranchialGraphStructure" | event branchial graph without labeling |
| "AllEventsBranchialGraph" | graph of branch pair event ancestry across all steps |
| "AllEventsBranchialGraphStructure" | all events branchial graph without labeling |
| "EvolutionEventBranchialGraph" | graph of evolution branch pair event ancestry at a given step |
| "EvolutionEventBranchialGraphStructure" | evolution event branchial graph without labeling |
| "AllEventsEvolutionBranchialGraph" | graph of evolution branch pair event ancestry across all steps |
| "AllEventsEvolutionBranchialGraphStructure" | all events evolution branchial graph without labeling |
| "BranchPairResolutionsList" | association of all resolved and unresolved branch pairs up to a given step |
| "EvolutionBranchPairResolutionsLIst" | association of all resolved and unresolved evolution branch pairs up to a given step |
| "CausalInvariantQ" | whether the system is causal invariant (all branch pairs converge) |
| "EvolutionCausalInvariantQ" | whether the system is evolution causal invariant (all evolution branch pairs converge) |
| "KnuthBendixCompletion" | list of Knuth–Bendix completion rules required to force causal invariance |
| "EvolutionKnuthBendixCompletion" | list of Knuth–Bendix completion rules required to force evolution causal invariance |
| "StateWeights" | list of weights for all vertices in the states graph |
| "PetriNetObjects" | list of PetriNetObject expressions obtained (default) |
| "LabeledGraphs" | list of directed graph representations obtained (with token counts represented graphically) |
| "LabeledGraphsHighlighted" | list of directed graph representations obtained (with token counts represented graphically, and with transition firings highlighted) |
| "WeightedGraphs" | list of directed graph representations obtained (with token counts represented as vertex weights) |
| "WeightedGraphsHighlighted" | list of directed graph representations obtained (with token counts represented as vertex weights, and with transition firings highlighted) |
| "Tokens" | list of token numbers associated to each place after each firing |
| "TokenFirings" | list of token numbers associated to each place after each firing, with the corresponding transition firing specified |
| "IncludeStepNumber" | False | whether to label states and events with their respective step numbers |
| "IncludeStateID" | False | whether to label states and events with unique IDs |
| "IncludeInitializationEvents" | False | whether to include pseudoevents that set up initial conditions |
| "IncludeEventInstances" | False | whether to show distinct updating events that connect the same states as separate edges |
| "IncludeStateWeights" | False | whether to weight state vertices by their rate of occurrence at a particular time step |
| "IncludeStatePathWeights" | False | whether to weight state vertices by the number of distinct evolution paths that lead to them |
| "StateRenderingFunction" | Automatic | how to label states that appear in graphs |
| "EventRenderingFunction" | Automatic | how to label events that appear in graphs |
| MaxItems | Infinity | how many instances of a causal graph or evolution causal graph to return |
| "GivePredecessors" | False | whether to label branch pairs with their predecessor state |
| "GiveResolvents" | False | whether to label branch pairs with their resolvent state |
| "IncludeSelfPairs" | False | whether to include trivial branch pairs |
| "IncludeFullBranchialSpace" | False | whether to show all possible states in a given branchial graph |
| "LineThickness" | 1 | absolute line thickness for graph edges |
| Inherited | use the explicit vertex name as the label |
| None | use no label for the vertex |
| "string" | use a shape from the VertexShapeFunction collection |
| func | apply the function func to the name of the vertex |
| "AssociationForm" | Petri net represented as an association of places, transitions and arcs |
| "Places" | list of places in the Petri net |
| "PlaceCount" | number of places in the Petri net |
| "Transitions" | list of transitions in the Petri net |
| "TransitionCount" | number of transitions in the Petri net |
| "Arcs" | list of arcs in the Petri net |
| "ArcCount" | number of arcs in the Petri net |
| "Tokens" | list of token numbers associated to each place in the Petri net |
| "TokenCounts" | total number of tokens across all places in the Petri net |
| "UnlabeledGraph" | directed graph form of the Petri net without token counts represented graphically |
| "LabeledGraph" | directed graph form of the Petri net with token counts represented graphically |
| "WeightedGraph" | directed graph form of the Petri net with token counts represented as vertex weights |
Construct a simple Petri net from lists of four places, three transitions and eight arcs, with an initial distribution of six tokens:
| In[1]:= |
| Out[1]= | 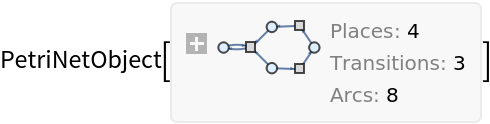 |
Showing basic multiway Petri net evolution:
| In[2]:= |
| Out[2]= | 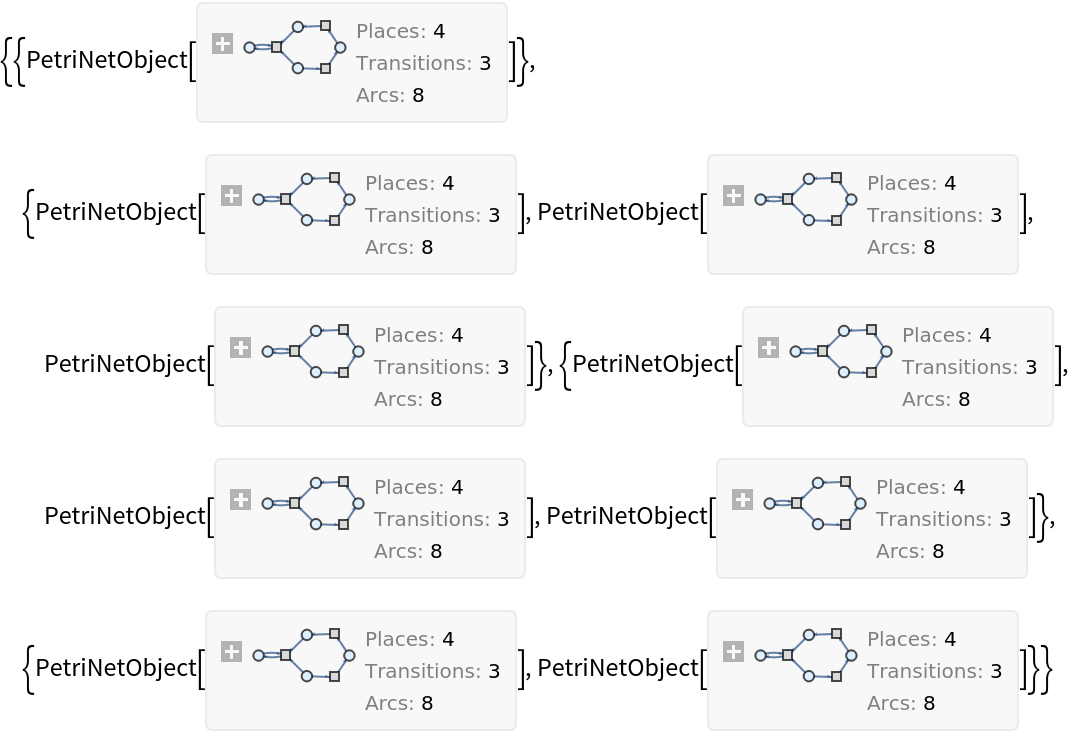 |
Show the sets of labeled graphs obtained after one step, with every possible transition firing highlighted:
| In[3]:= |
| Out[3]= | 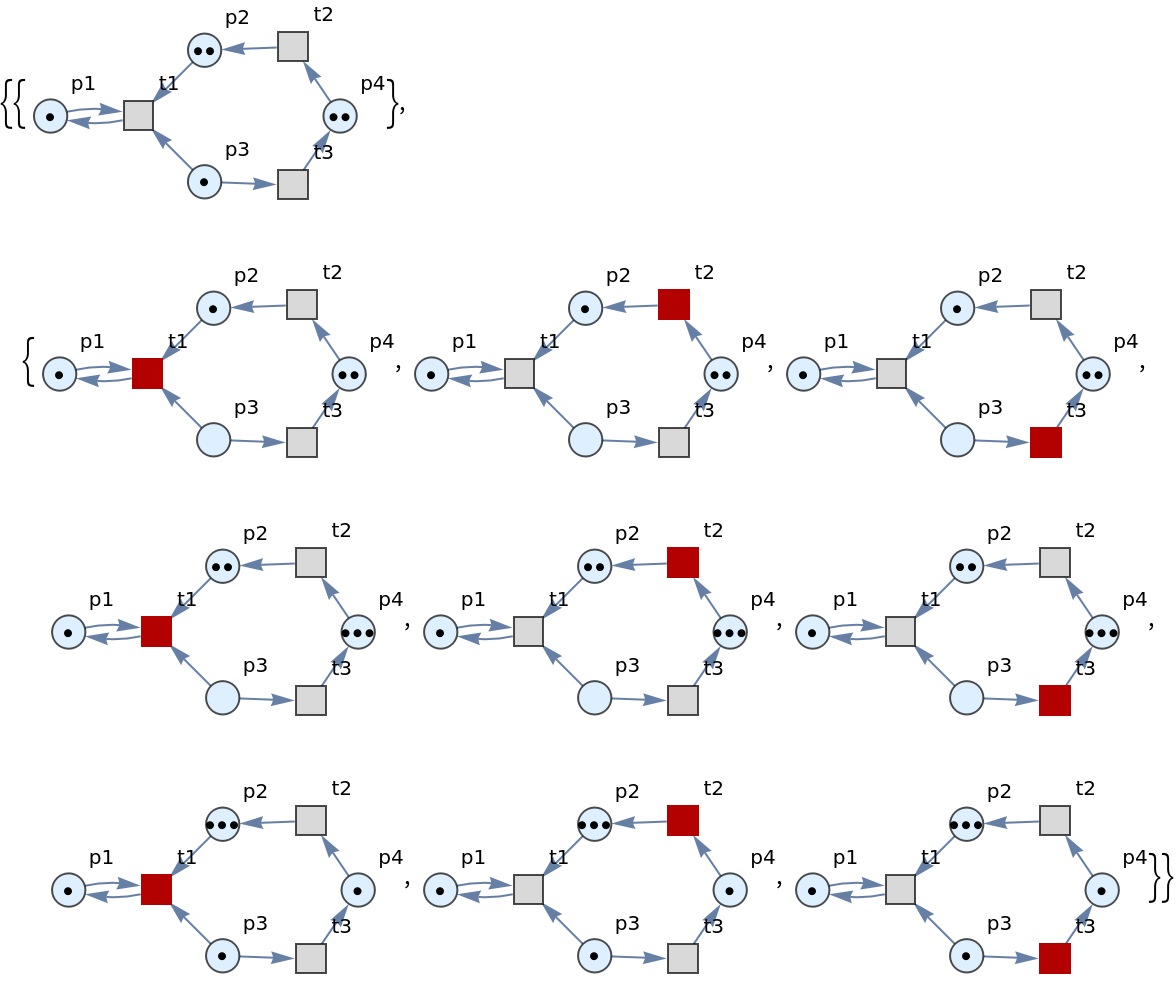 |
Show the sets of labeled graphs after three steps, without any transition firings highlighted:
| In[4]:= |
| Out[4]= | 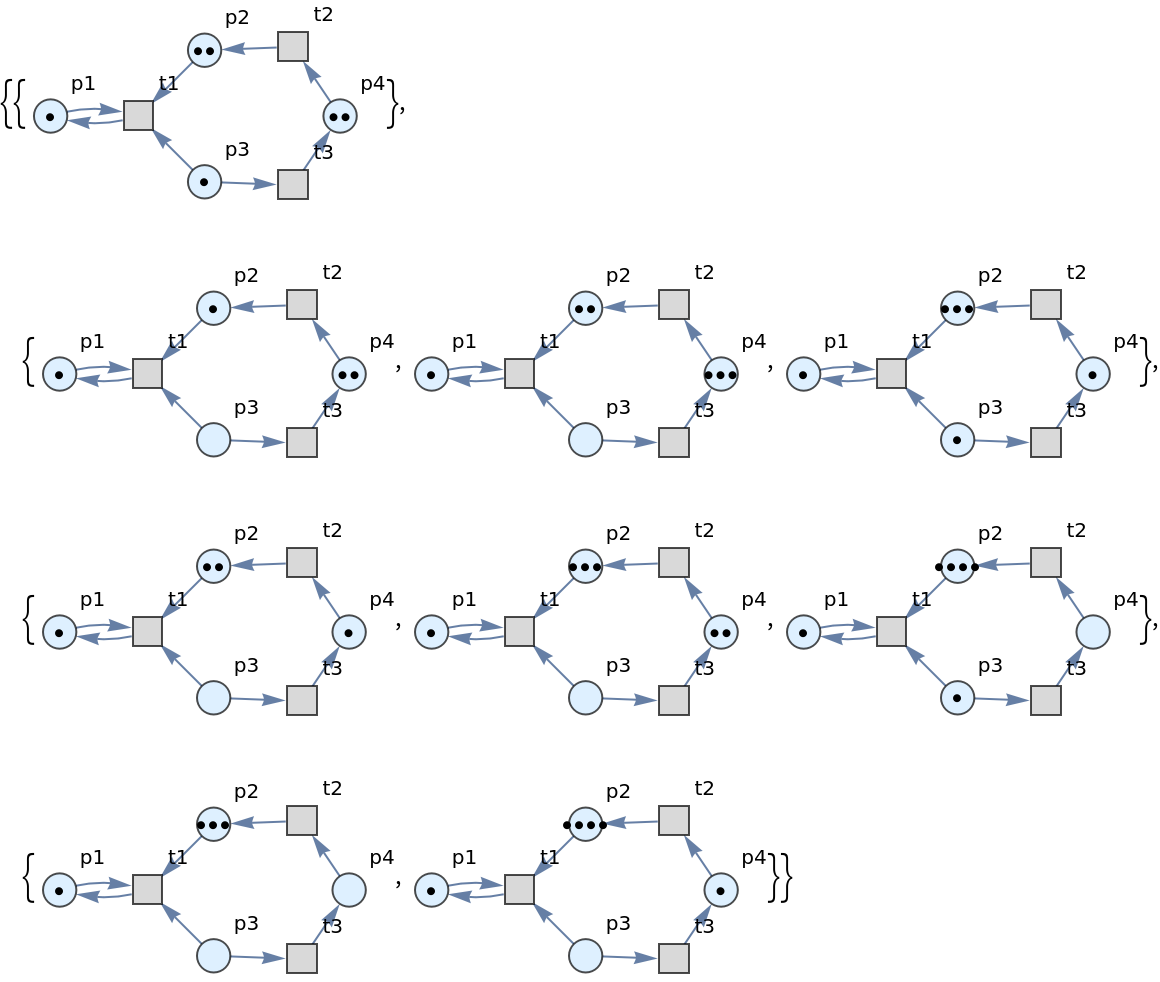 |
Generate a graph showing how each state is obtained from the others:
| In[5]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 3, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/43d8d536faff083f.png) |
| Out[5]= | 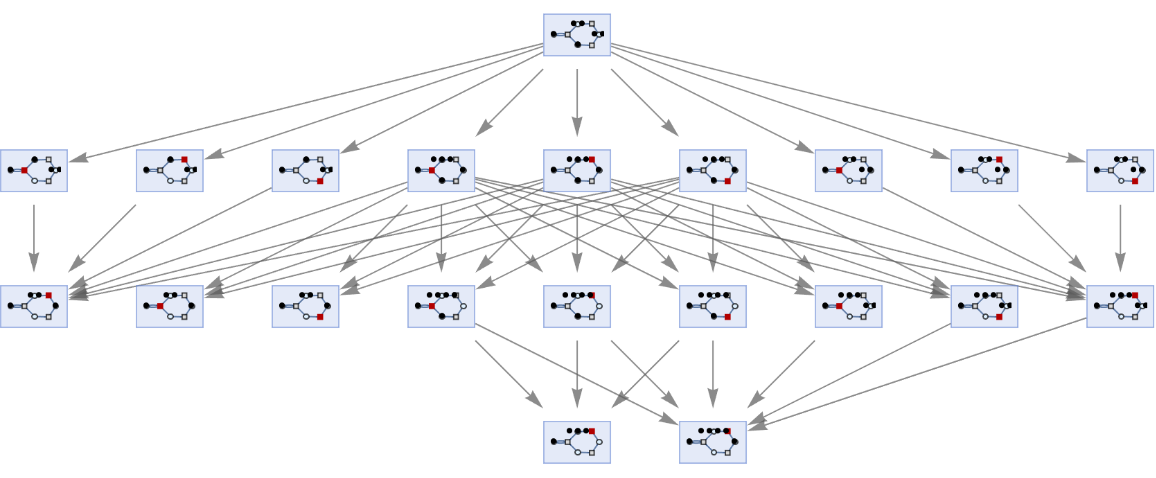 |
Show the structure of the graph, without labels:
| In[6]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 3, "StatesGraphStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/331f6707f06094dd.png) |
| Out[6]= |  |
Generate the list of updating events applied at each step:
| In[7]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, p4}, "Transitions" -> {t1, t2, t3}, "Arcs" -> {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}|>, {1, 2, 1, 2}, 2, "AllEventsList"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/37901ccb7d210429.png) |
| Out[7]= | 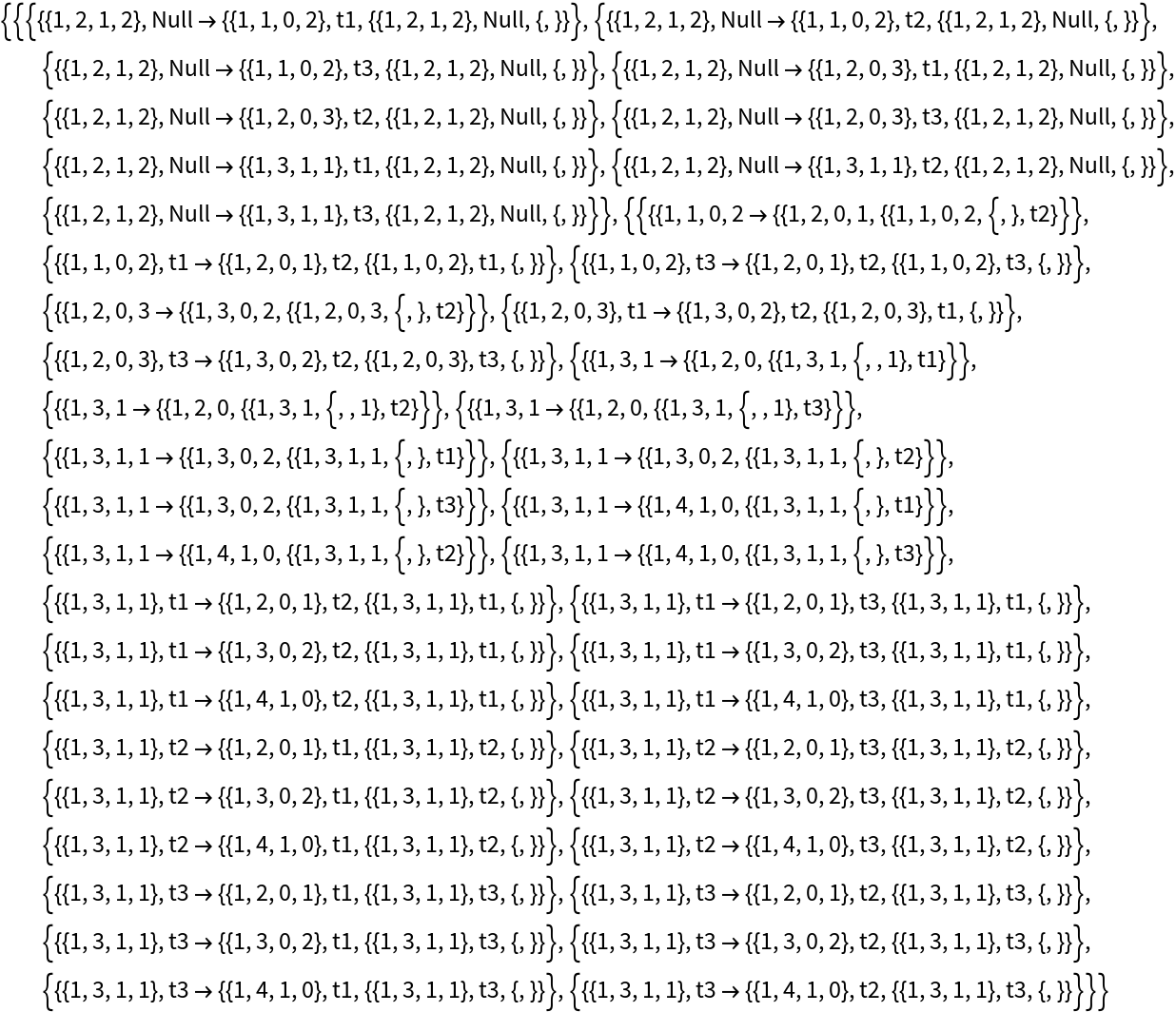 |
Construct a Petri net representing a reversible chemical reaction, specified by an association of three places, two transitions and six arcs, with an initial distribution of 12 tokens:
| In[8]:= | ![petriNet = ResourceFunction["MakePetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/2532297ef7629961.png) |
| Out[8]= |  |
Generate a graph of the evolution history, with updating events included:
| In[9]:= |
| Out[9]= | 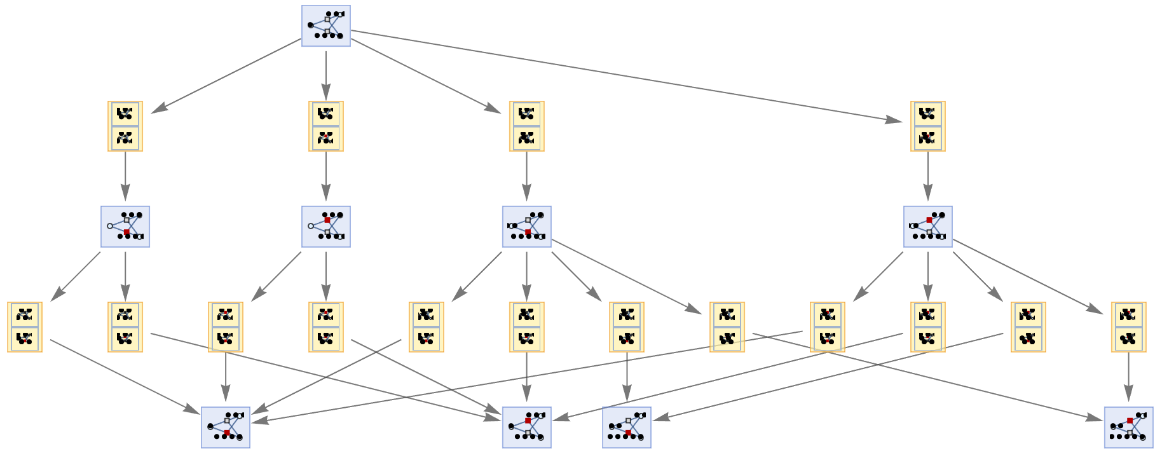 |
Show the structure of the graph, without labels:
| In[10]:= |
| Out[10]= | 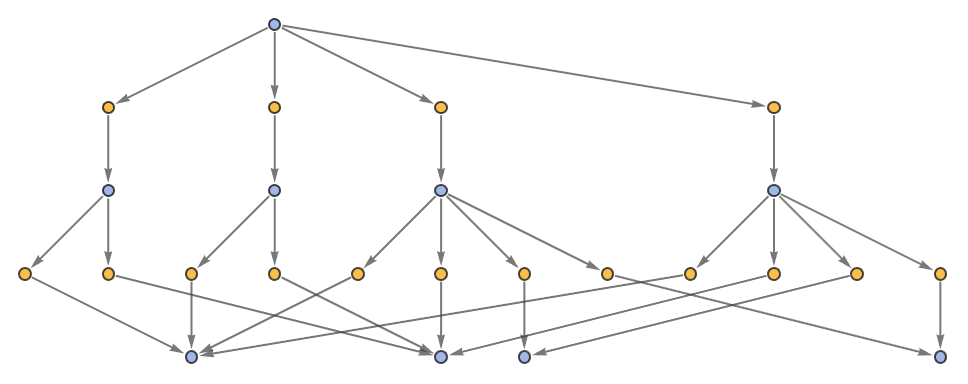 |
Generate the causal graph, showing dependencies between updating events:
| In[11]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "CausalGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/6501915f20c032a6.png) |
| Out[11]= | 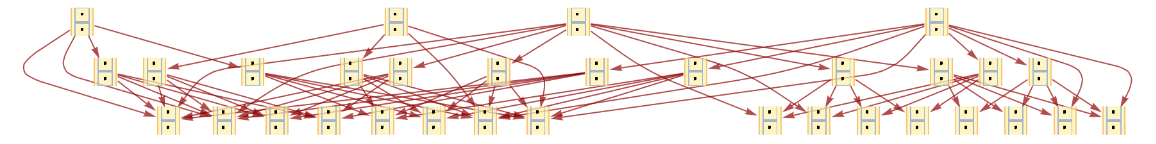 |
Show the structure of the graph, without labels:
| In[12]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "CausalGraphStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/3b055db998344435.png) |
| Out[12]= | 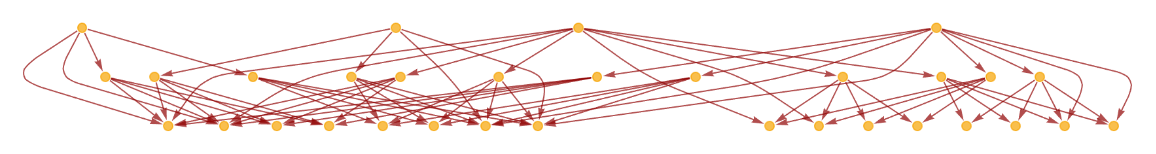 |
Generate the evolution events graph, with causal connections included:
| In[13]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "EvolutionCausalGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1a3ce3b4d7107035.png) |
| Out[13]= | 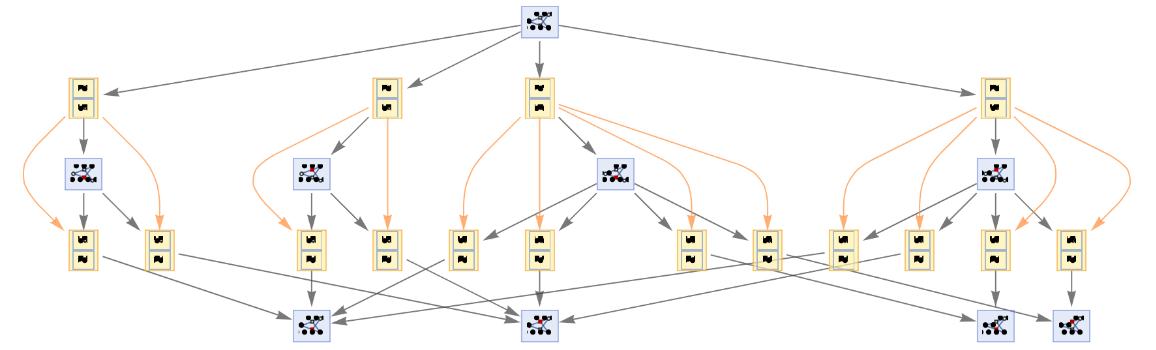 |
Show the structure of the graph, without labels:
| In[14]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "EvolutionCausalGraphStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/07e12c3b94ad0eb3.png) |
| Out[14]= | 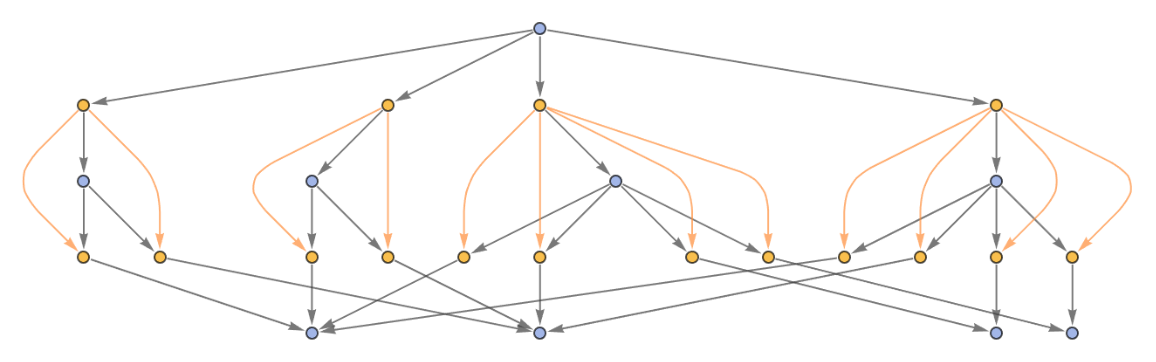 |
Specify an event selection function that picks a random event at each step:
| In[15]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|> -> "Random", {1, 4, 7}, 4, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/3fddb54264d858fb.png) |
| Out[15]= | 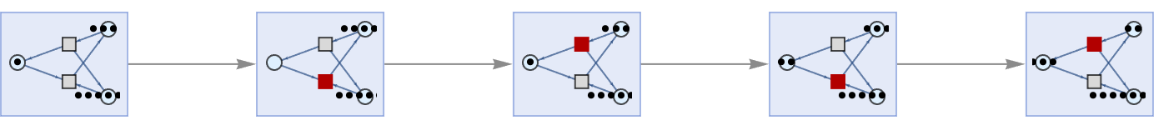 |
Generate causal graphs for all possible choices of event sequences:
| In[16]:= | ![Graph[#, VertexSize -> 1] & /@ Take[ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}, {1, 4, 7}, 3, "CausalGraphInstances"], 10]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/558f2b50778bc0b3.png) |
| Out[16]= | 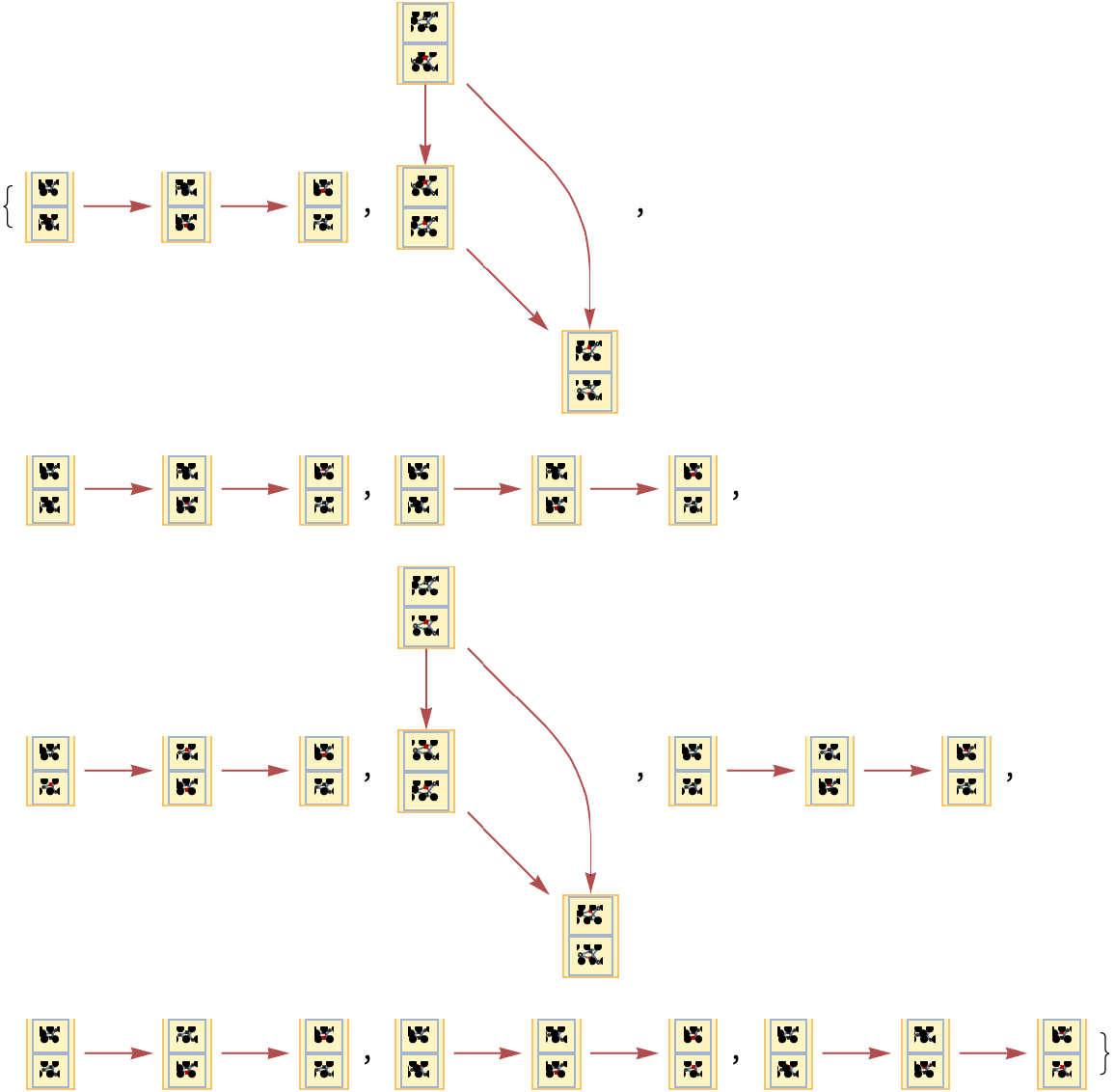 |
Show the structures of the graphs, without labels:
| In[17]:= | ![Take[ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}, {1, 4, 7}, 3, "CausalGraphStructureInstances"], 10]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/031e7f122155a379.png) |
| Out[17]= | 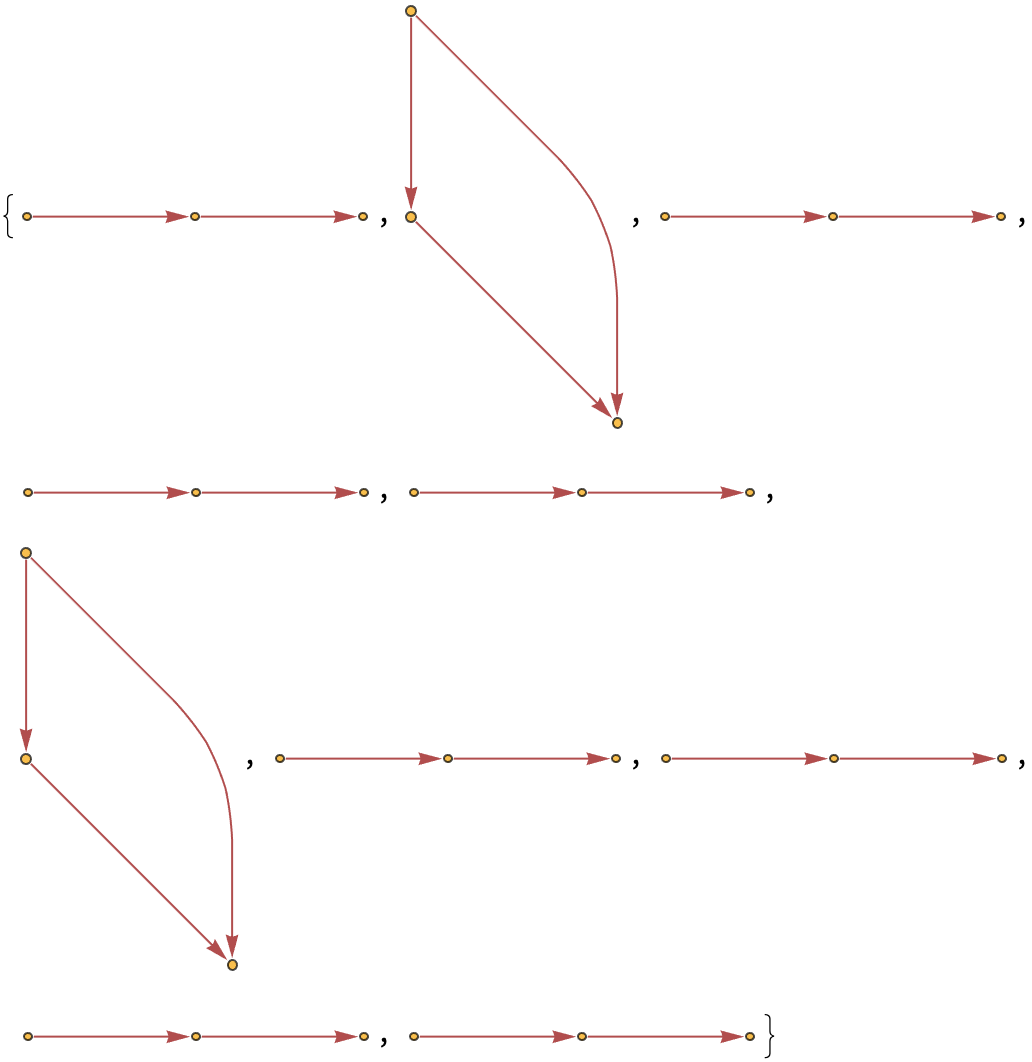 |
Generate the list of all branch pairs (i.e. critical pairs):
| In[18]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 3, "BranchPairsList"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/12d95fe2e17a442a.png) |
| Out[18]= |  |
Generate the association showing which branch pairs converged and which did not:
| In[19]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 3, "BranchPairResolutionsList"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0f6999699622d747.png) |
| Out[19]= |  |
Prove that the system is causal invariant:
| In[20]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 3, "CausalInvariantQ"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/44754168b2f196ce.png) |
| Out[20]= |
Construct a Petri net representing a simple producer-consumer problem in concurrency theory:
| In[21]:= | ![petriNet = ResourceFunction["MakePetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/065d290c1cb40e43.png) |
| Out[21]= | 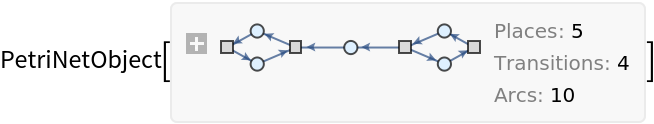 |
Generate a graph showing branch pair ancestry:
| In[22]:= |
| Out[22]= | 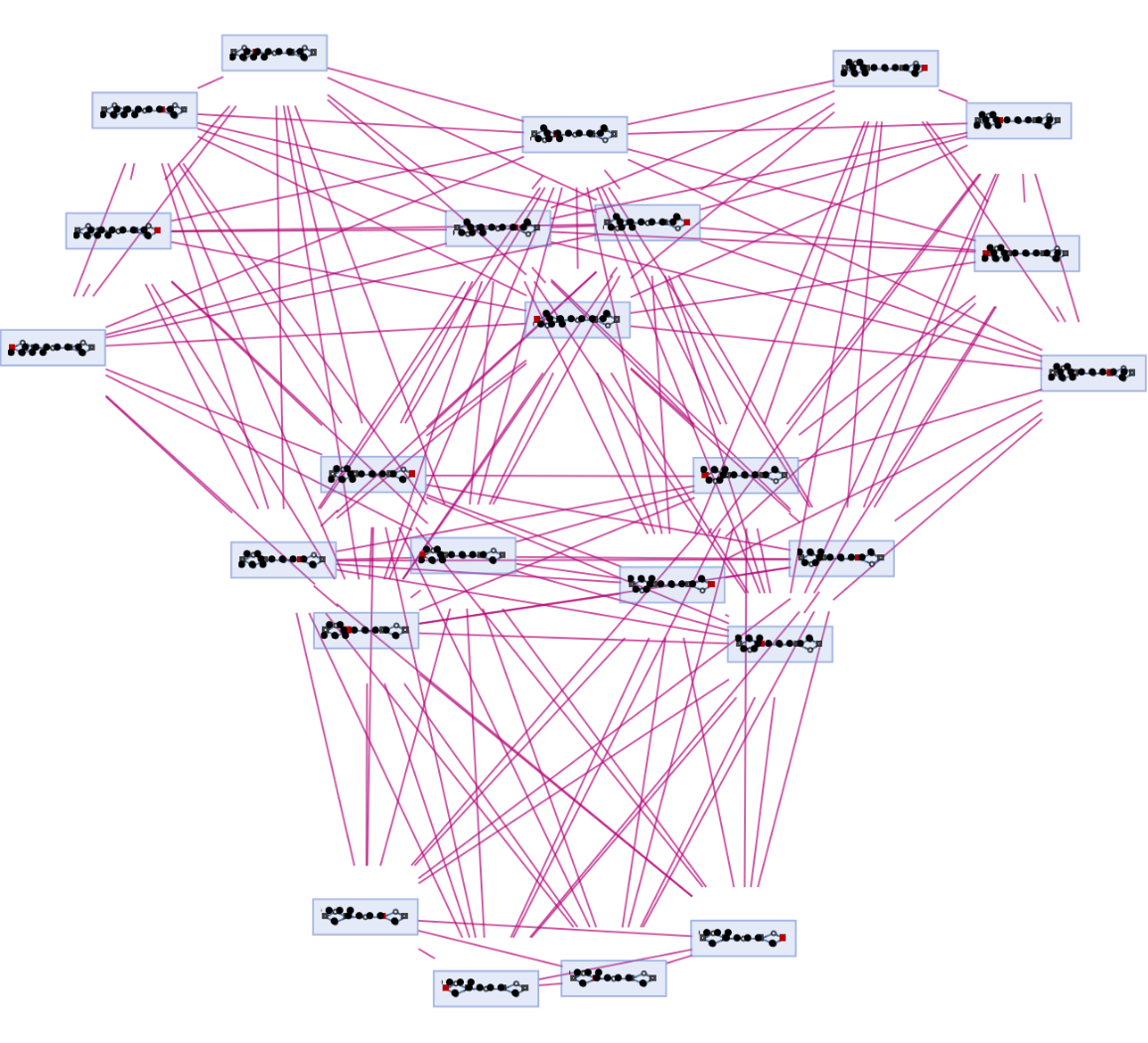 |
Show the structure of the graph, without labels:
| In[23]:= |
| Out[23]= | 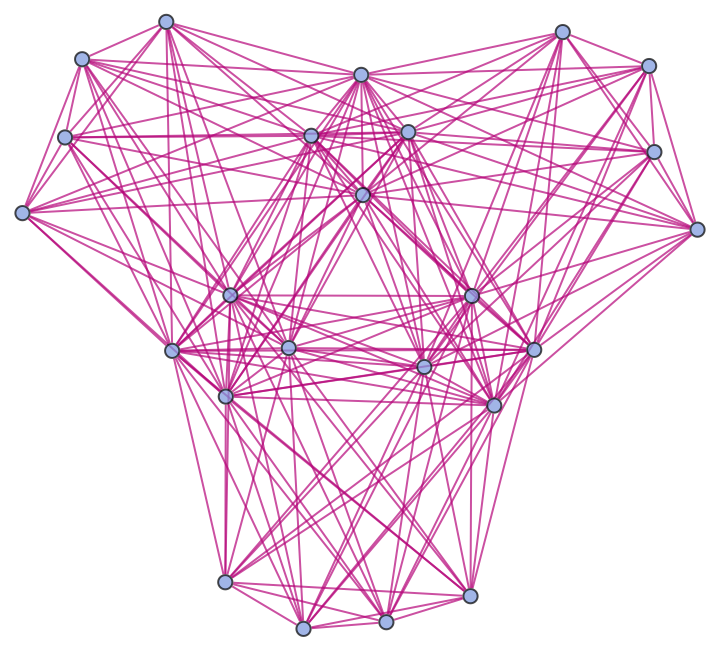 |
Generate a graph showing branch pair event ancestry:
| In[24]:= |
| Out[24]= | 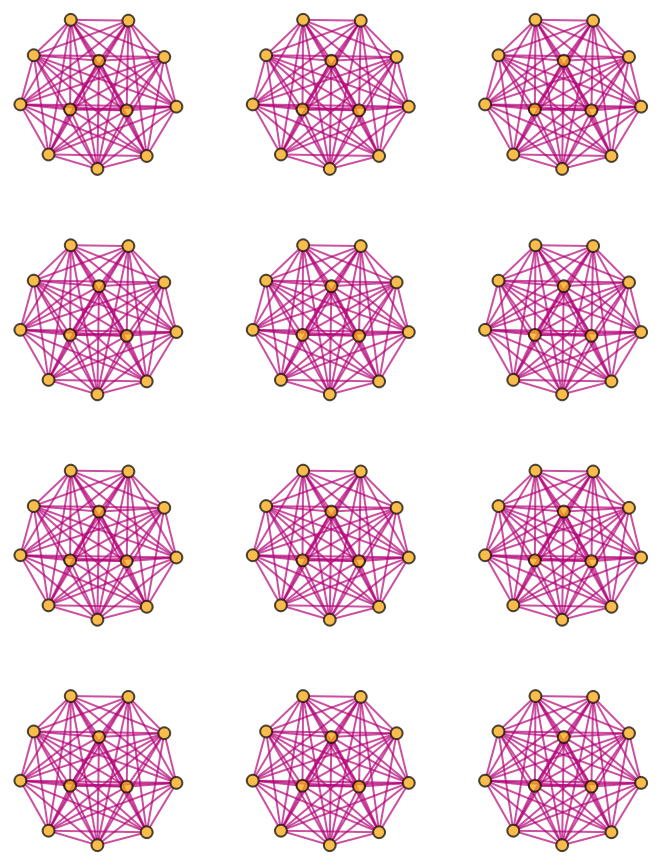 |
Prevent identical states from being merged by including step numbers and state IDs:
| In[25]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 2, "StatesGraph", "IncludeStepNumber" -> True, "IncludeStateID" -> True, VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1a11a2505370231e.png) |
| Out[25]= | 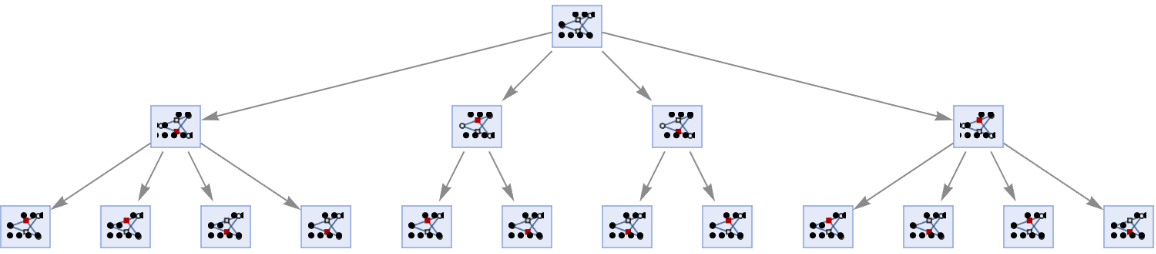 |
| In[26]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 2, "AllStatesList", "IncludeStepNumber" -> True, "IncludeStateID" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1a02c26eee7f6b8b.png) |
| Out[26]= |  |
Generate a graph of full evolution history, with all events included:
| In[27]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}, 2, "EvolutionGraphFullStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/26a2d250a7ebb17d.png) |
| Out[27]= | 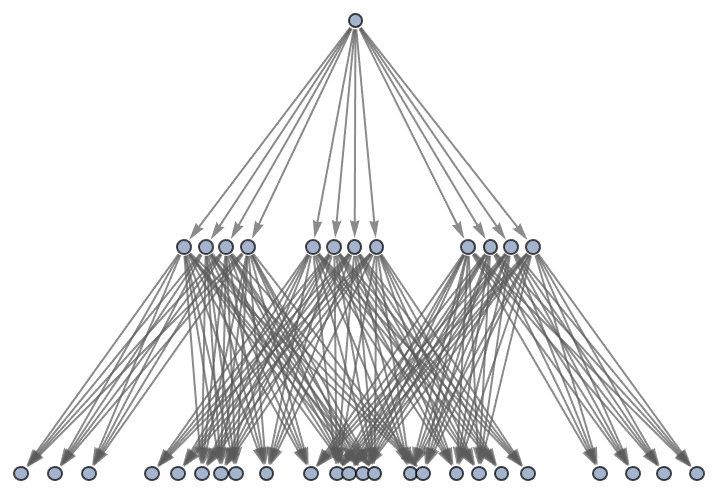 |
Generate a graph of full evolution history, with no merging of equivalent states:
| In[28]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}, 2, "EvolutionGraphUnmergedStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/6209023462bd309b.png) |
| Out[28]= | 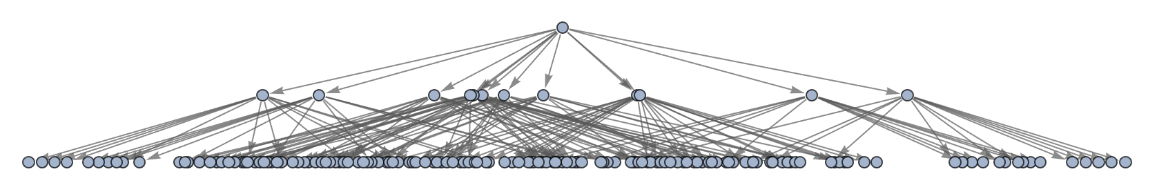 |
Generate a graph of evolution history, with edges weighted by event multiplicity:
| In[29]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}, 3, "EvolutionGraphWeightedStructure", EdgeLabels -> "EdgeWeight"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/267ae45633658f8d.png) |
| Out[29]= |  |
Generate a states graph with vertices weighted by their rate of occurrence on each time step:
| In[30]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, "Buffer", p4}, "Transitions" -> {"Produce", t1, t2, "Consume"}, "Arcs" -> {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}|>, {1, 3, 0, 6, 2}, 2, "StatesGraph", "IncludeStateWeights" -> True, VertexSize -> 2, VertexLabels -> "VertexWeight", AspectRatio -> 1/2]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0a45937ea6e8d0c3.png) |
| Out[30]= | 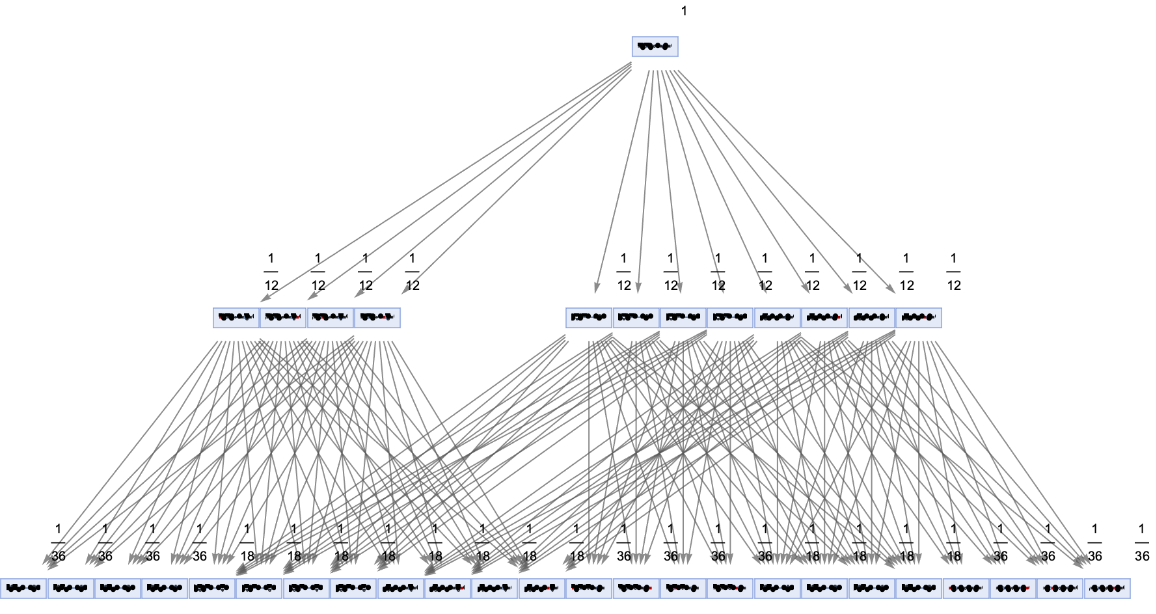 |
Show the structure of the graph, without labels:
| In[31]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, "Buffer", 4}, "Transitions" -> {"Produce", t1, t2, "Consume"}, "Arcs" -> {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}|>, {1, 3, 0, 6, 2}, 2, "StatesGraphStructure", "IncludeStateWeights" -> True, VertexLabels -> "VertexWeight", AspectRatio -> 1/2]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/4b4c11f6e3215af1.png) |
| Out[31]= | 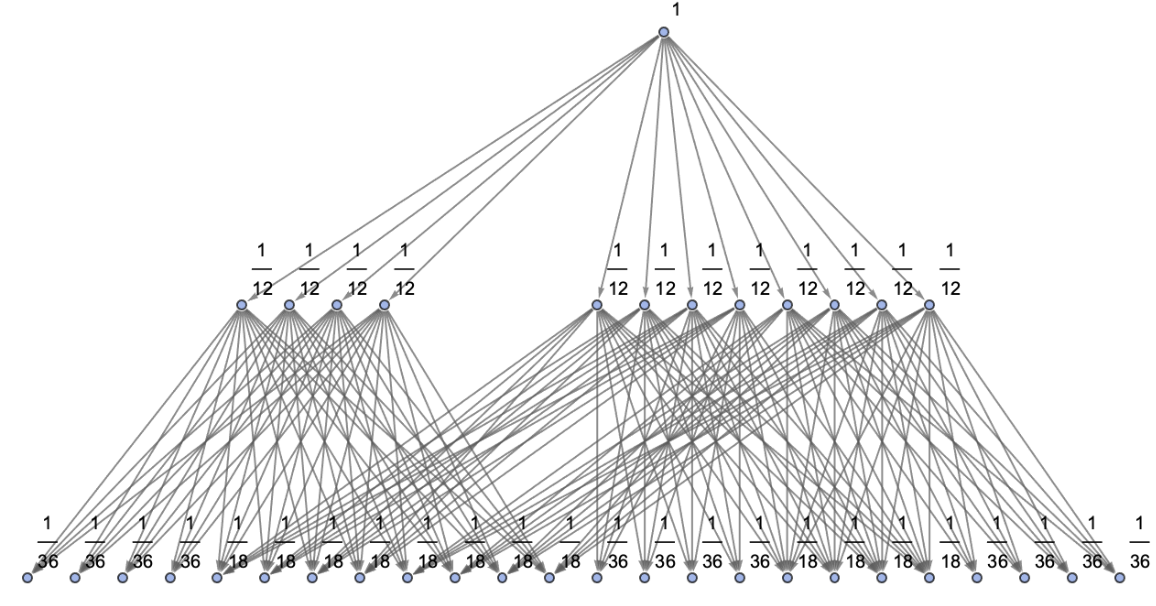 |
Generate a states graph with vertices weighted by the number of distinct evolution paths that lead to them:
| In[32]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}, 2, "StatesGraph", "IncludeStatePathWeights" -> True, VertexSize -> 2, VertexLabels -> "VertexWeight", AspectRatio -> 1/2]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/51fafc7ad402b35c.png) |
| Out[32]= | 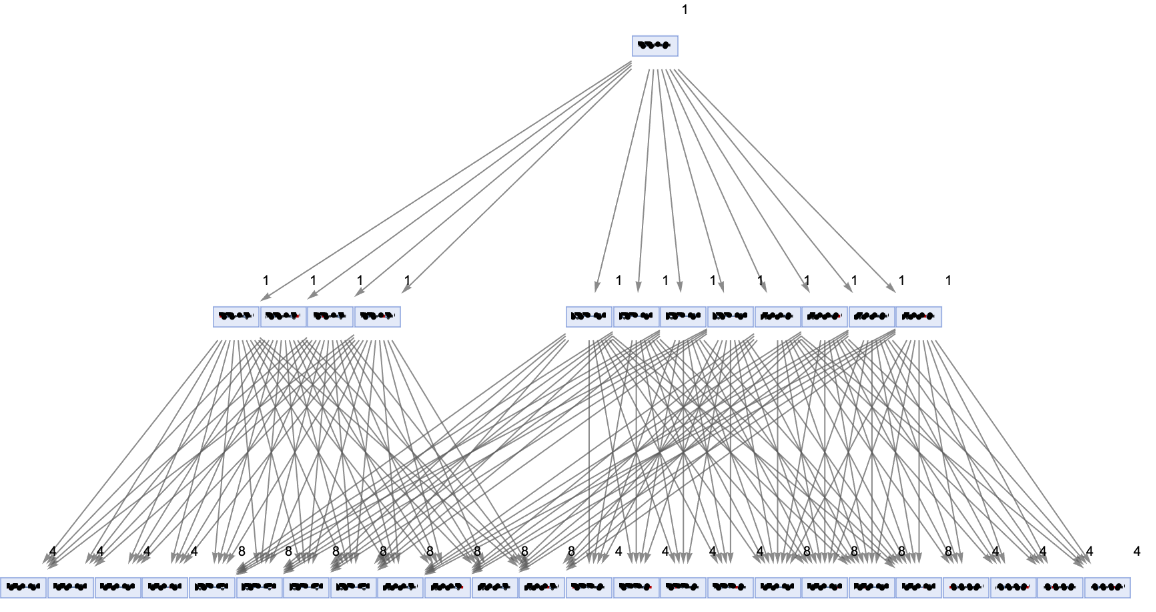 |
Show the structure of the graph, without labels:
| In[33]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}, 2, "StatesGraphStructure", "IncludeStatePathWeights" -> True, VertexLabels -> "VertexWeight", AspectRatio -> 1/2]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/29fc5eee9b364a16.png) |
| Out[33]= | 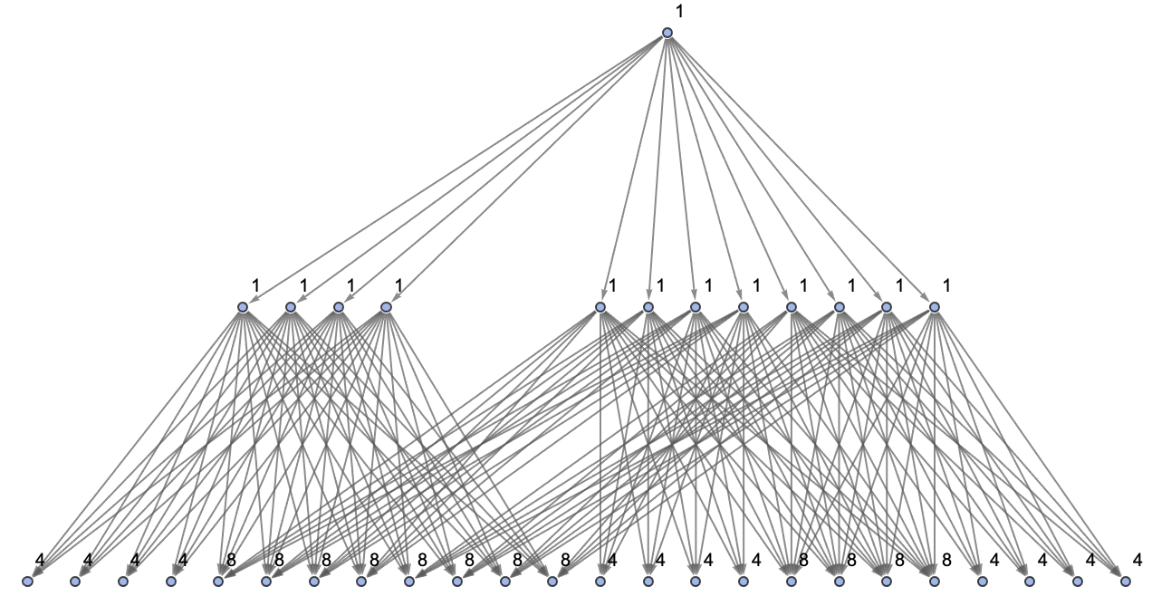 |
MultiwayPetriNet accepts PetriNetObject expressions:
| In[34]:= | ![ResourceFunction["MultiwayPetriNet"][
ResourceFunction["MakePetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {1, 1, 1}], 2, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1f9a370cc8c6f025.png) |
| Out[34]= | 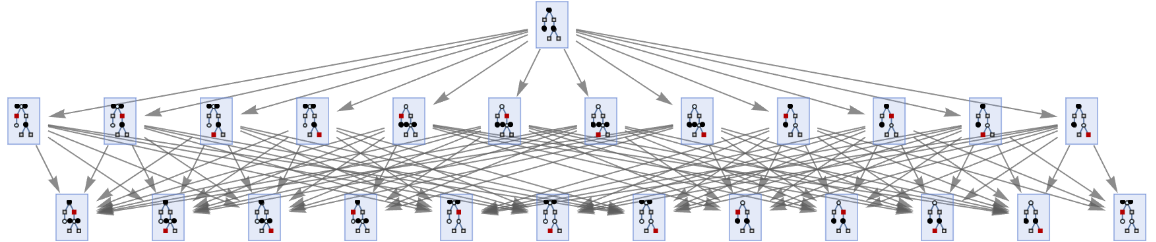 |
Or lists of places, transitions and arcs:
| In[35]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {2, 0, 0}, 2, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/41faa1bedabe1049.png) |
| Out[35]= | 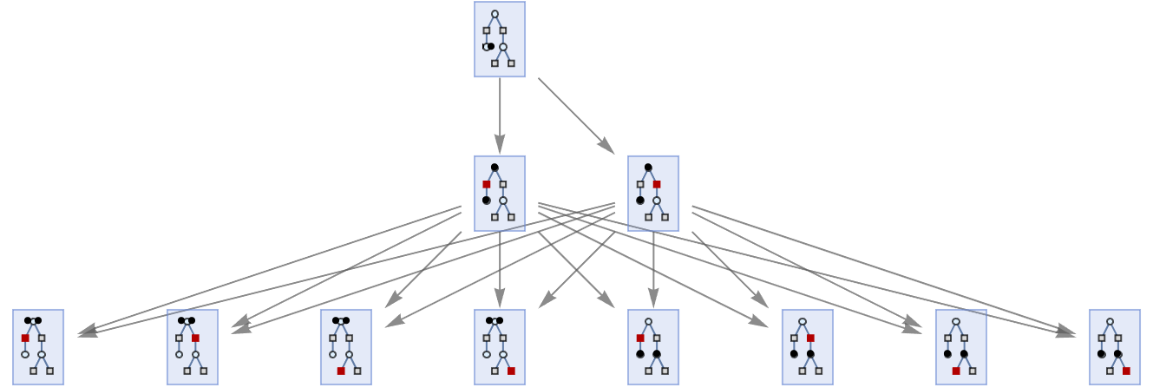 |
Or associations of places, transitions and arcs:
| In[36]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3}, "Transitions" -> {t1, t2, t3, t4}, "Arcs" -> {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}|>, {0, 2, 0}, 2, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/3ab52ad67f6ebcc4.png) |
| Out[36]= | 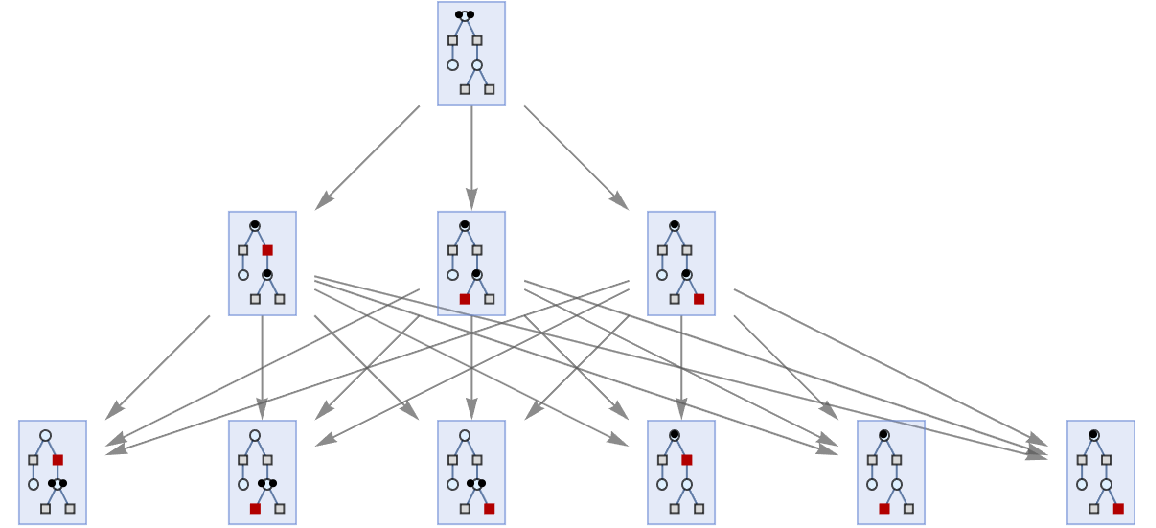 |
If MultiwayPetriNet is called without an explicit list of token numbers, it is assumed that each place contains no tokens:
| In[37]:= |
| Out[37]= |
Apply only the first possible event at each step:
| In[38]:= | ![ResourceFunction["MultiwayPetriNet"][
ResourceFunction["MakePetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {1, 1, 1}] -> "Sequential", 5, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1b2b37f822b6c303.png) |
| Out[38]= |  |
Apply the first and last possible events at each step:
| In[39]:= | ![ResourceFunction["MultiwayPetriNet"][
ResourceFunction["MakePetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {1, 1, 1}] -> ({First[#], Last[#]} &), 3, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/6f585191850979dd.png) |
| Out[39]= | 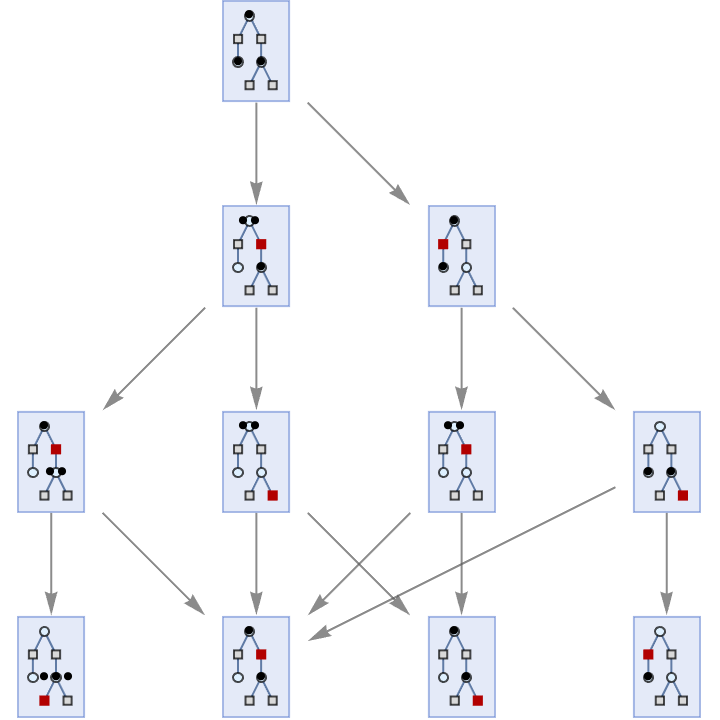 |
Apply an event selection function for a Petri net specified in list form:
| In[40]:= | ![ResourceFunction[
"MultiwayPetriNet"][{{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}} -> ({First[#], Last[#]} &), {2, 0, 0}, 3, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/4109b2672457dcf4.png) |
| Out[40]= | 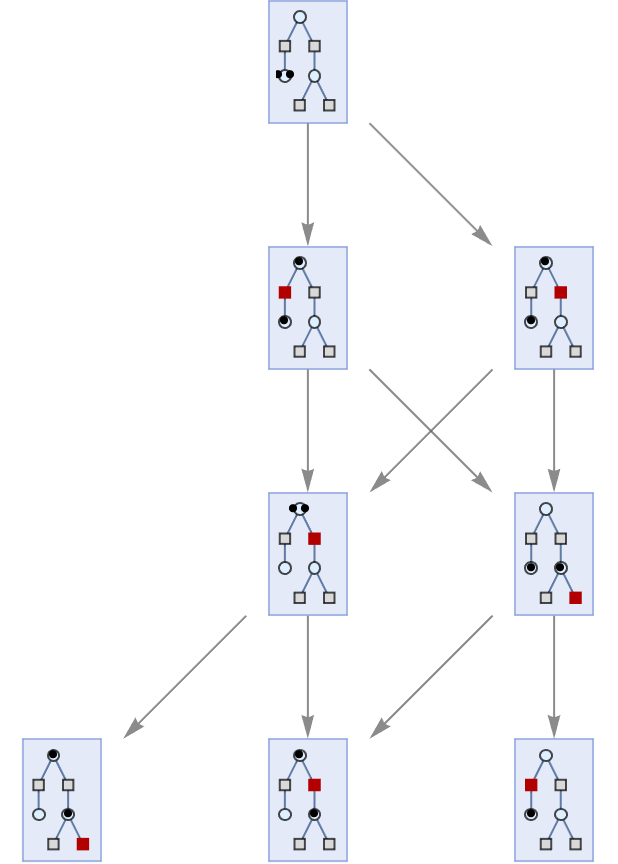 |
Apply an event selection function for a Petri net specified in association form:
| In[41]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3}, "Transitions" -> {t1, t2, t3, t4}, "Arcs" -> {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}|> -> ({First[#], Last[#]} &), {0, 2, 0}, 3, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/04d9167562ecf278.png) |
| Out[41]= | 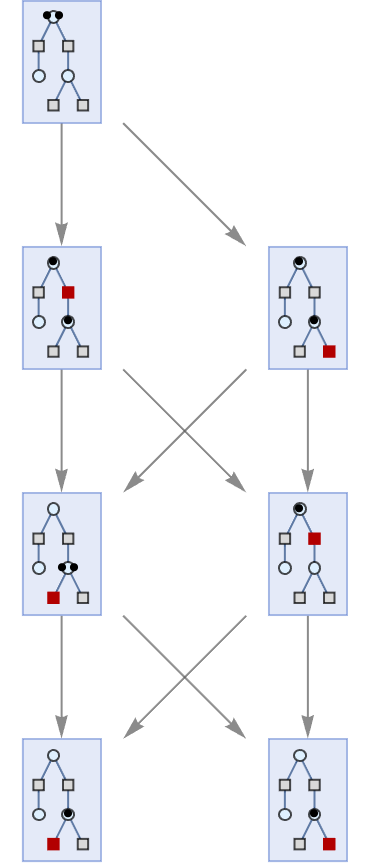 |
Construct a Petri net representing a simple producer-consumer problem in concurrency theory:
| In[42]:= | ![petriNet = ResourceFunction["MakePetriNet"][{p1, p2, p3, "Buffer", p4}, {"Produce", t1, t2, "Consume"}, {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}, {1, 3, 0, 6, 2}]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/08c08e1dbad7aa36.png) |
| Out[42]= | 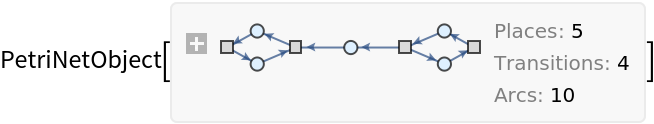 |
Show the list of Petri net objects obtained after two steps of multiway evolution:
| In[43]:= |
| Out[43]= | 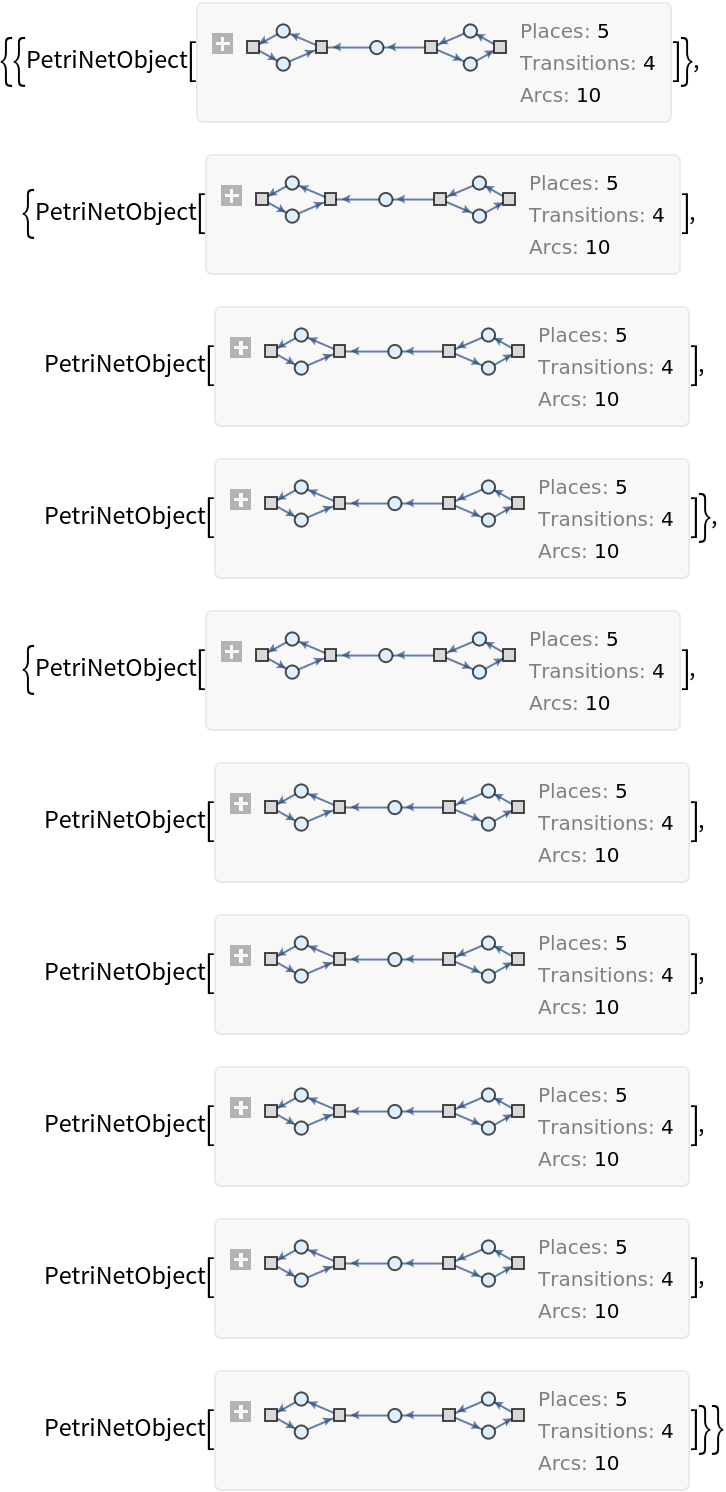 |
Show the list of directed graph representations (with token counts represented graphically) after two steps of multiway evolution:
| In[44]:= |
| Out[44]= | 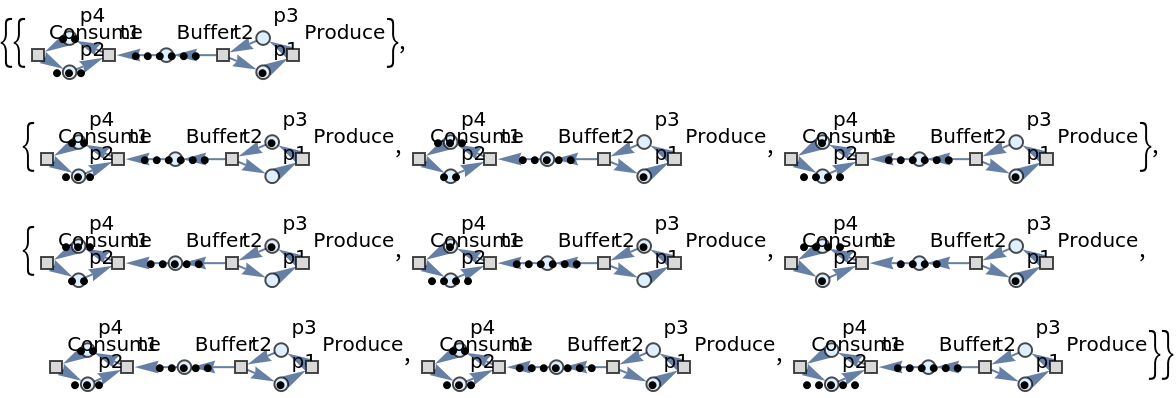 |
Show the list of directed graph representations (with token counts represented graphically, and with transition firings highlighted) after two steps of multiway evolution:
| In[45]:= |
| Out[45]= | 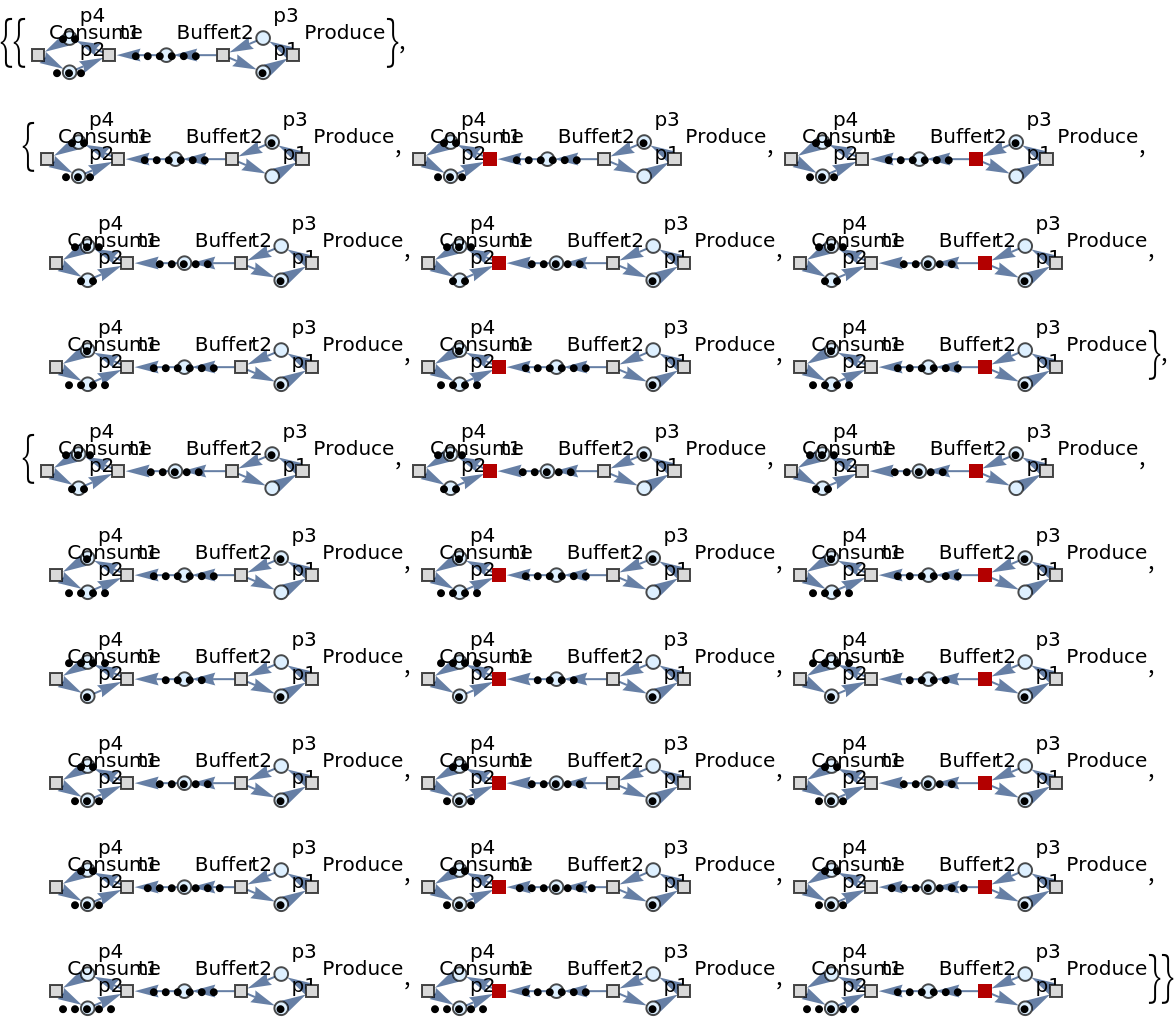 |
Show the list of directed graph representations (with token counts represented as vertex weights) after two steps of multiway evolution:
| In[46]:= |
| Out[46]= | 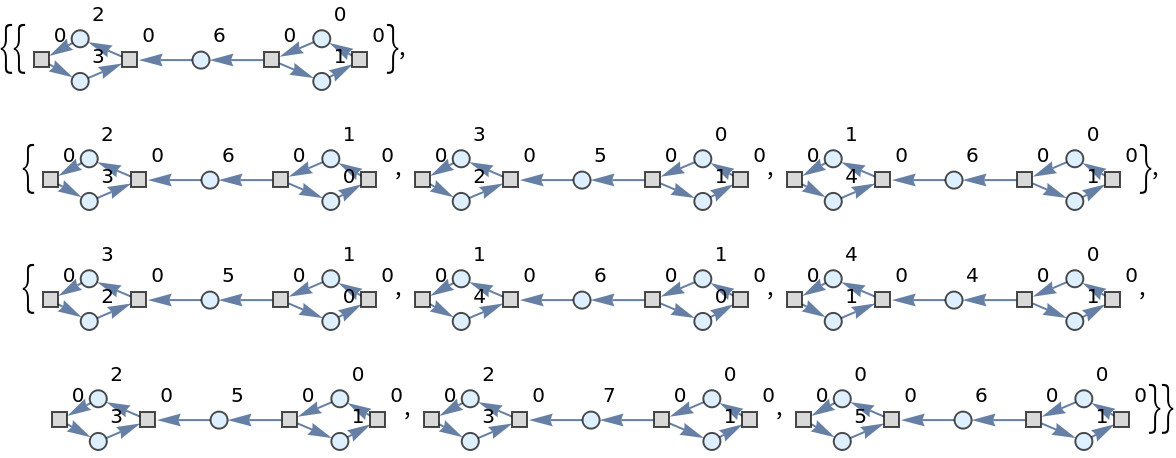 |
Show the list of directed graph representations (with token counts represented as vertex weights, and with transition firings highlighted) after two steps of multiway evolution:
| In[47]:= |
| Out[47]= | 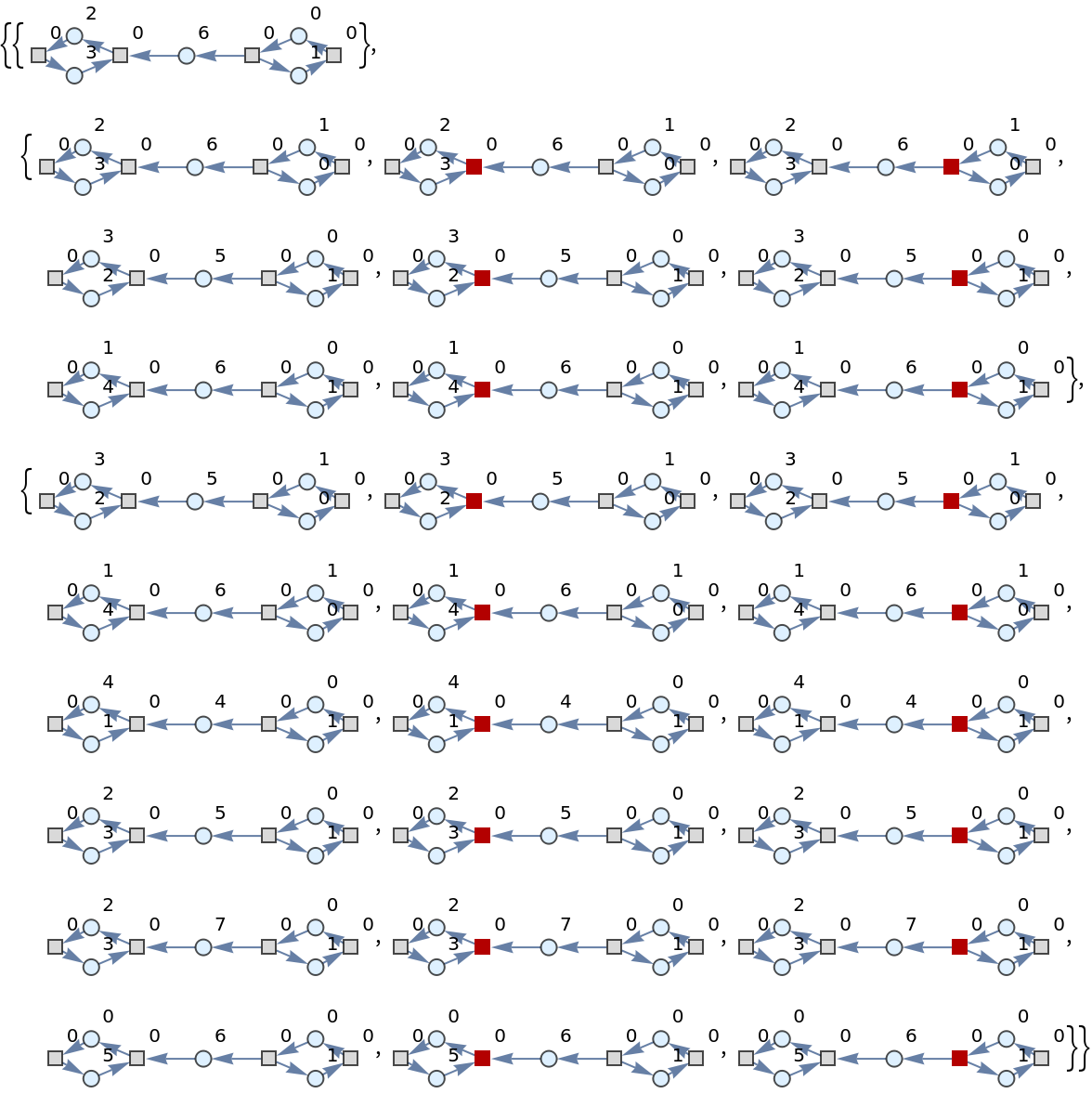 |
Show the list of token numbers associated to each place after each firing, following two steps of multiway evolution:
| In[48]:= |
| Out[48]= |
Show the list of token numbers associated to each place after each firing (with the corresponding transition firings specified), following two steps of multiway evolution:
| In[49]:= |
| Out[49]= |  |
By default, states are labeled by their contents:
| In[50]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 2, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/15e3829b9c1af927.png) |
| Out[50]= | 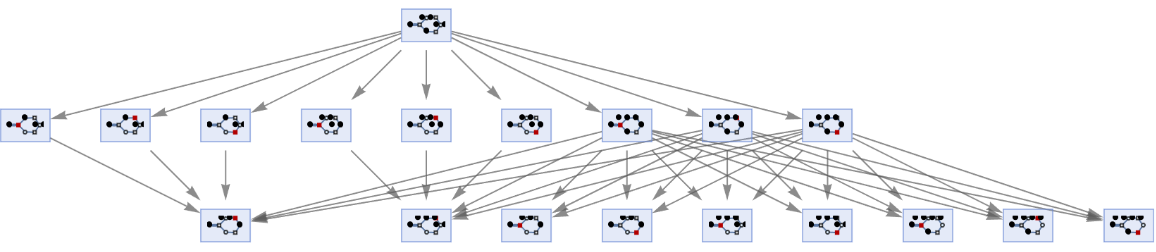 |
Use no labeling for states:
| In[51]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 2, "StatesGraph", "StateRenderingFunction" -> None]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/114616a05e599124.png) |
| Out[51]= | 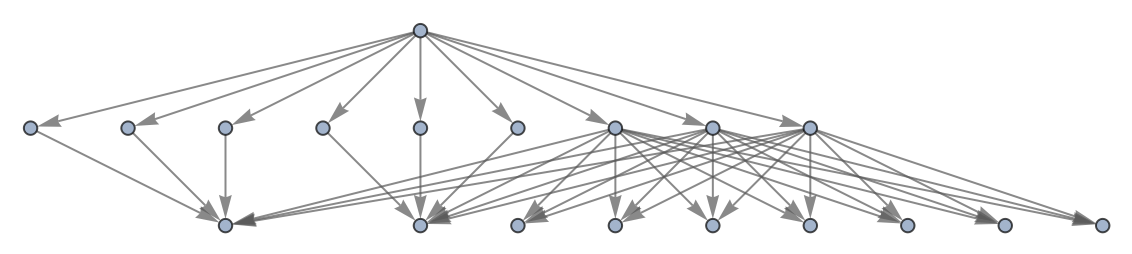 |
"StatesGraphStructure" yields the same result:
| In[52]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 2, "StatesGraphStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/29f6d1dc721f1057.png) |
| Out[52]= | 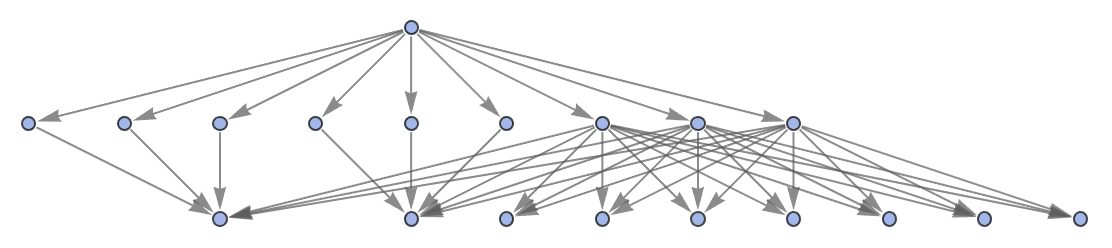 |
Use raw state names as node labels:
| In[53]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 2, "StatesGraph", "StateRenderingFunction" -> Inherited]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1180932663068fff.png) |
| Out[53]= |  |
Use a named shape as each state label:
| In[54]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, 2, "StatesGraph", "StateRenderingFunction" -> "Square"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0e67095de57443d8.png) |
| Out[54]= | 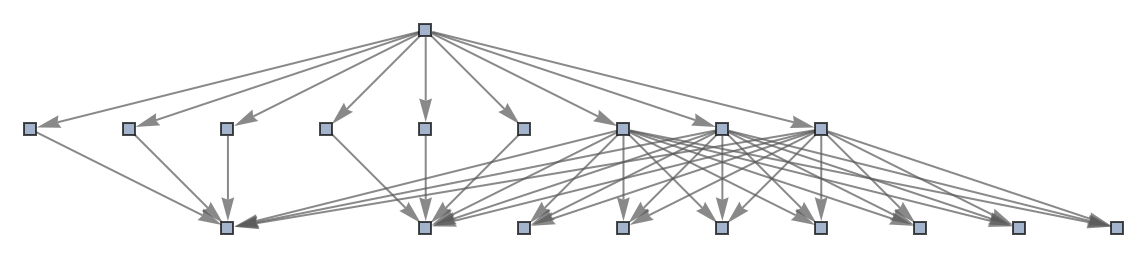 |
By default, both states and events are labeled by their contents:
| In[55]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "EvolutionEventsGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1611ab71c83909eb.png) |
| Out[55]= | 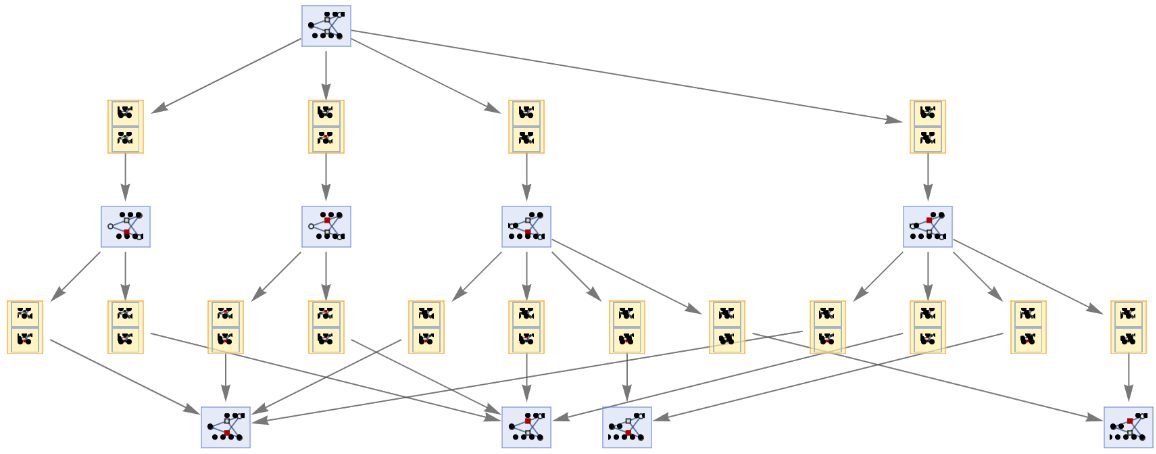 |
Use no labels for states or events:
| In[56]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "EvolutionEventsGraph", "StateRenderingFunction" -> None, "EventRenderingFunction" -> None]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/062350785fe66f56.png) |
| Out[56]= | 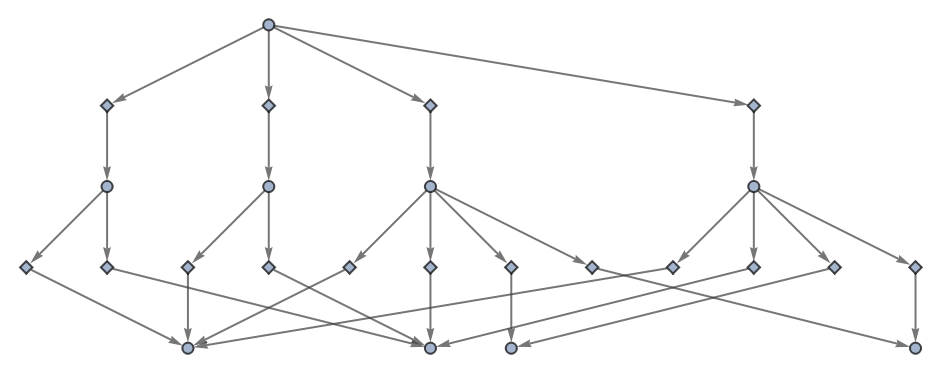 |
"EvolutionEventsGraphStructure" yields an equivalent result:
| In[57]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "EvolutionEventsGraphStructure"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/25501f13230e159c.png) |
| Out[57]= | 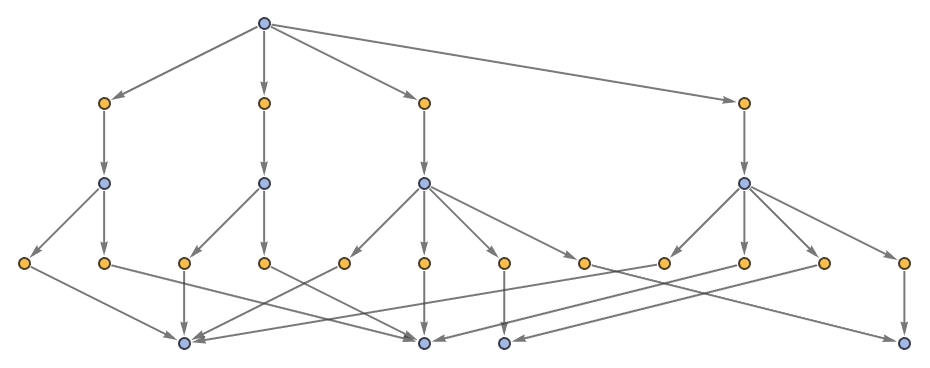 |
Use raw event expressions as their labels:
| In[58]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "EvolutionEventsGraph", "StateRenderingFunction" -> None, "EventRenderingFunction" -> Inherited]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/578cd1f1e2f0c91b.png) |
| Out[58]= | 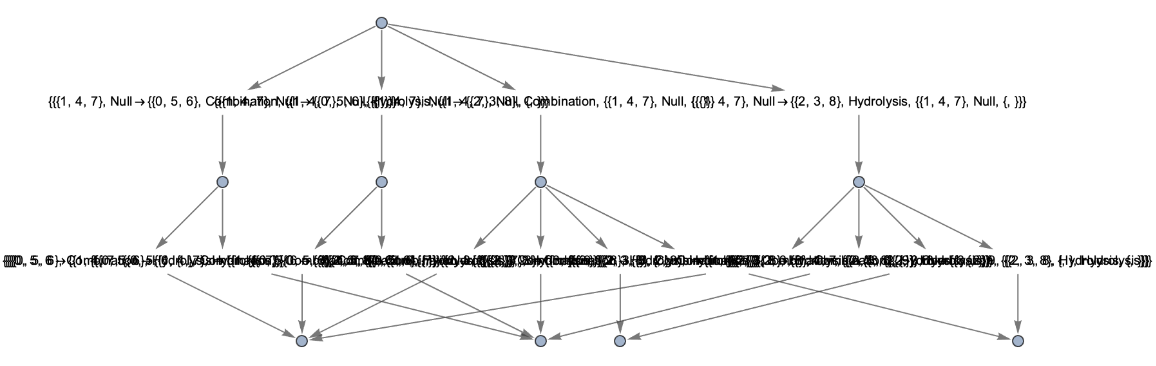 |
By default, "AllEventsList" does not include initialization events:
| In[59]:= |
| Out[59]= |  |
The option "IncludeInitializationEvents" allows one to override this default:
| In[60]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {2, 0, 0}, 2, "AllEventsList", "IncludeInitializationEvents" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/657fb525ffc36fe7.png) |
| Out[60]= | 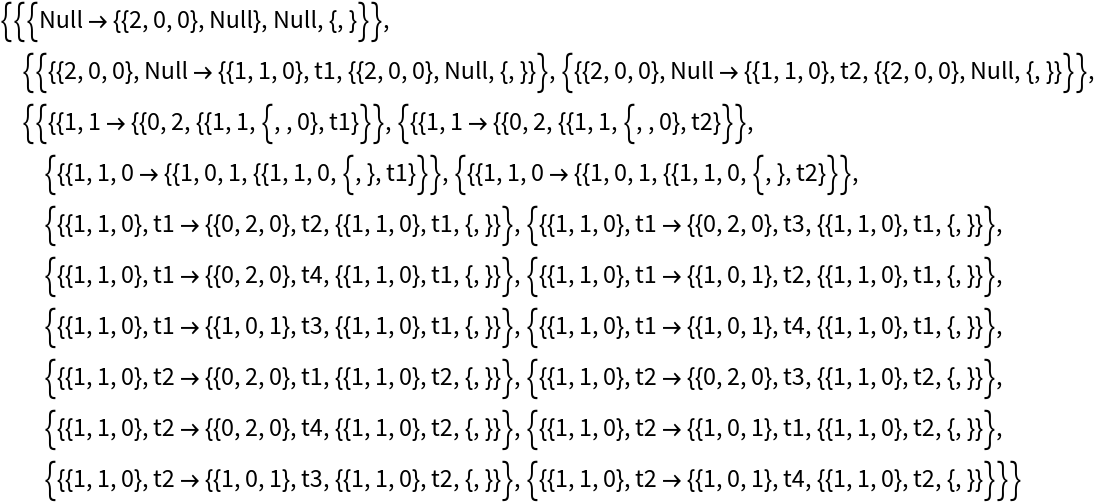 |
Initialization events have special default rendering:
| In[61]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {2, 0, 0}, 2, "EvolutionEventsGraph", "IncludeInitializationEvents" -> True, VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/1b6818267817ecf3.png) |
| Out[61]= | 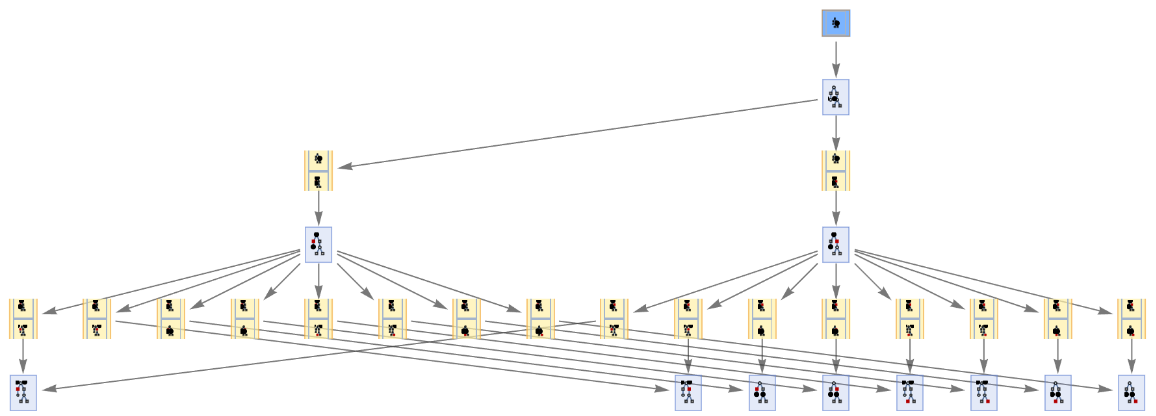 |
Place arrows in the middle of edges:
| In[62]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {2, 0, 0}, 2, "StatesGraph", EdgeShapeFunction -> GraphElementData["ShortFilledArrow", "ArrowSize" -> 0.03], VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/68bccd1fd0792b38.png) |
| Out[62]= | 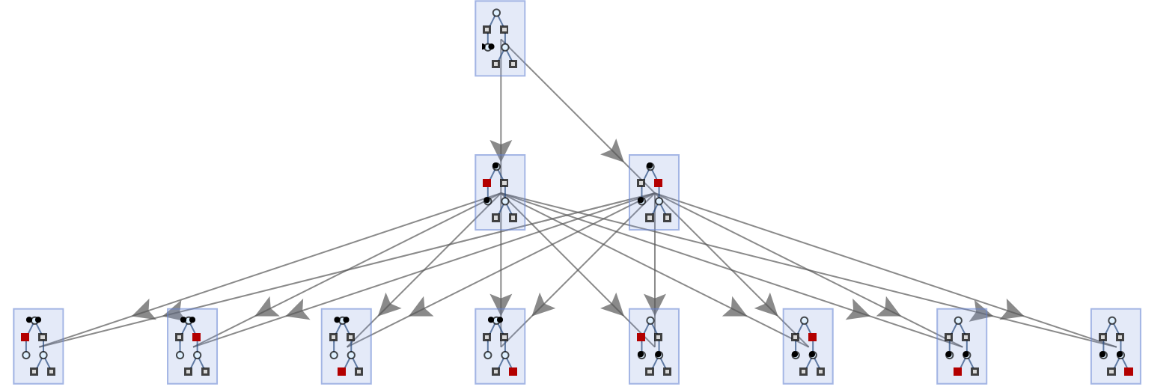 |
Generate an example of multiway Petri net evolution:
| In[63]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, "Buffer", p4}, "Transitions" -> {"Produce", t1, t2, "Consume"}, "Arcs" -> {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}|>, {1, 3, 0, 6, 2}, 3, "StatesGraph", VertexSize -> 3]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/67c98b165c3dbfd3.png) |
| Out[63]= | 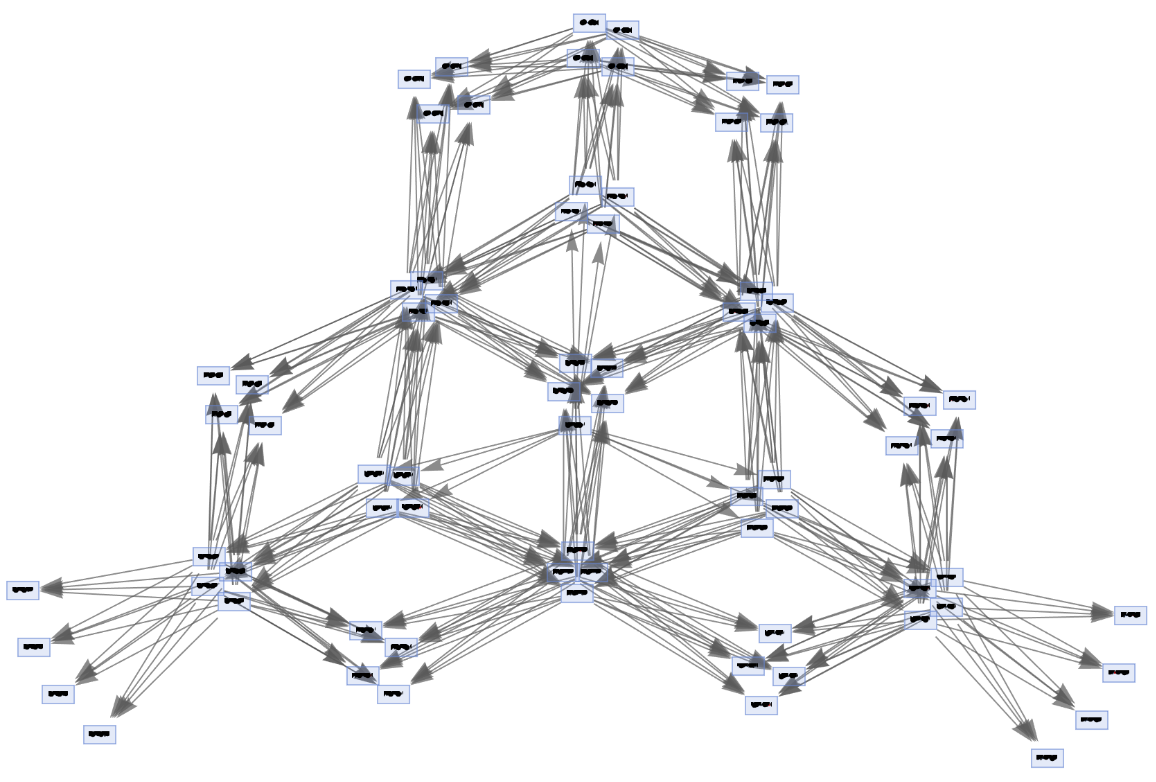 |
Force a layered digraph embedding:
| In[64]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, "Buffer", p4}, "Transitions" -> {"Produce", t1, t2, "Consume"}, "Arcs" -> {p1 -> "Produce", "Produce" -> p3, p3 -> t2, t2 -> p1, t2 -> "Buffer", "Buffer" -> t1, t1 -> p4, p4 -> "Consume", "Consume" -> p2, p2 -> t1}|>, {1, 3, 0, 6, 2}, 3, "StatesGraph", GraphLayout -> "LayeredDigraphEmbedding", AspectRatio -> 1/2, VertexSize -> 2]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/13c6c7950c4d56ea.png) |
| Out[64]= | 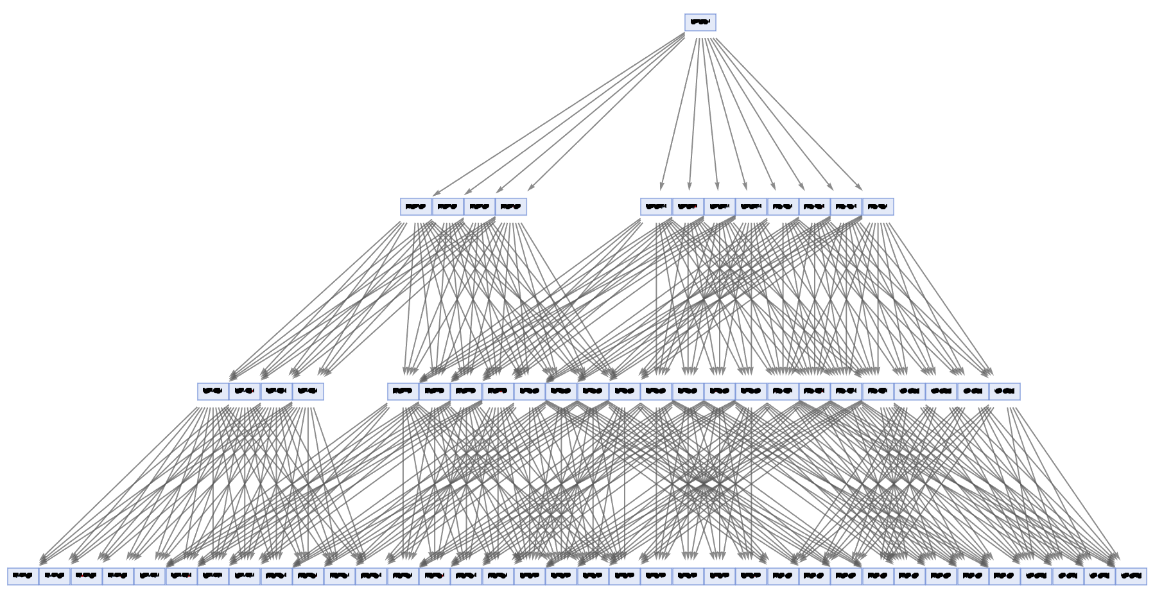 |
By default, equivalent states are merged across all time steps:
| In[65]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "StatesGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/081c24c3f59657ec.png) |
| Out[65]= | 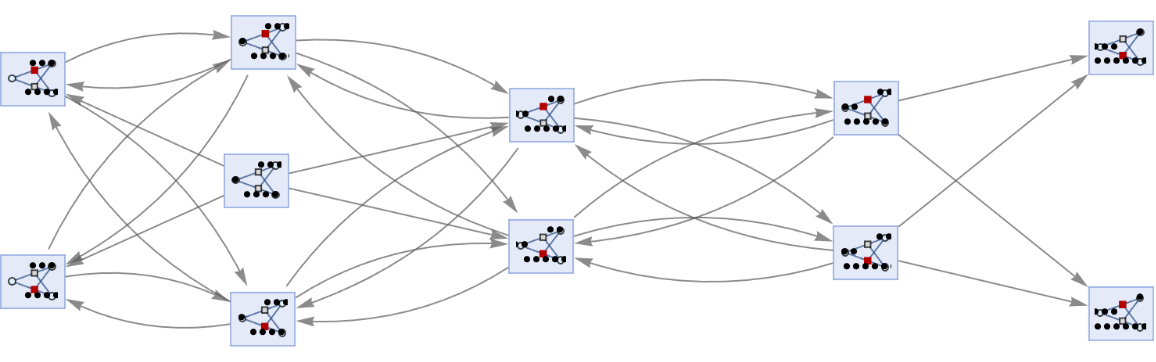 |
| In[66]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "AllStatesList"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/539407669e9557e9.png) |
| Out[66]= |  |
Merging of equivalent states across different time steps can be prevented by including step numbers:
| In[67]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "StatesGraph", "IncludeStepNumber" -> True, VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/2c4e5d8bc5d5ccb6.png) |
| Out[67]= | 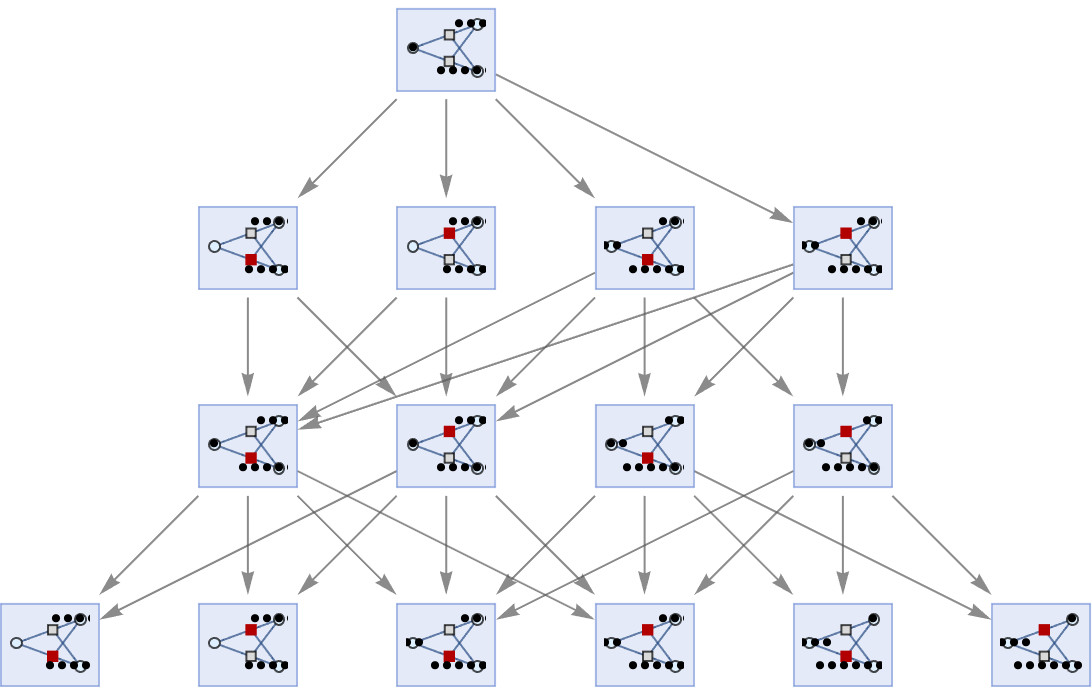 |
| In[68]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "AllStatesList", "IncludeStepNumber" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/2c2c1b3bfd8bc5ee.png) |
| Out[68]= |  |
Merging of equivalent states at the same time step can be prevented by also including state IDs:
| In[69]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "StatesGraph", "IncludeStepNumber" -> True, "IncludeStateID" -> True, VertexSize -> 3]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/32ec9e216a836ebc.png) |
| Out[69]= |  |
| In[70]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "AllStatesList", "IncludeStepNumber" -> True, "IncludeStateID" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/76334da701b6af42.png) |
| Out[70]= |  |
Step numbers and IDs also apply to events:
| In[71]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "EvolutionEventsGraph", "IncludeStepNumber" -> True, "IncludeStateID" -> True, VertexSize -> 3]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0bbdf377464df7bc.png) |
| Out[71]= | 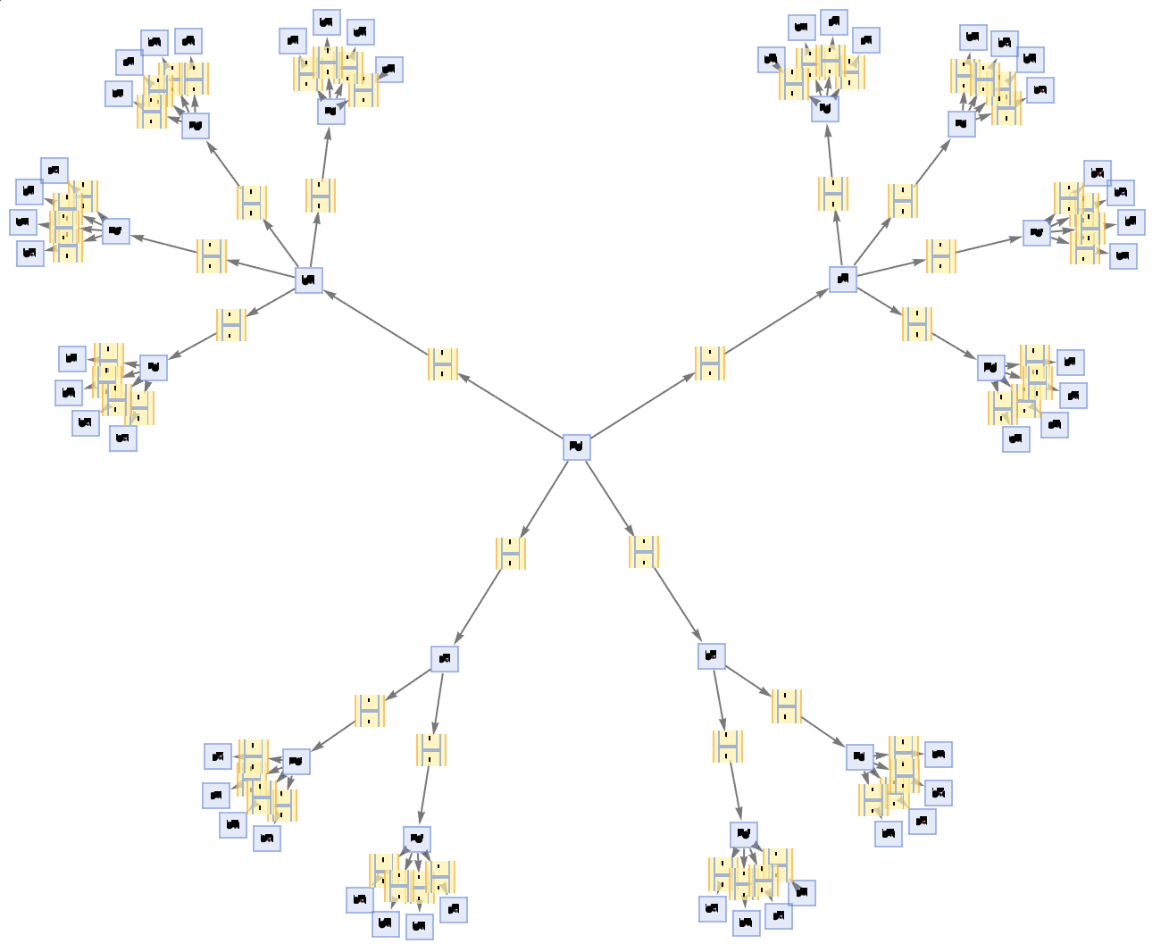 |
See the events:
| In[72]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 2, "AllEventsList", "IncludeStepNumber" -> True, "IncludeStateID" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/4d6aec7d6de48688.png) |
| Out[72]= | 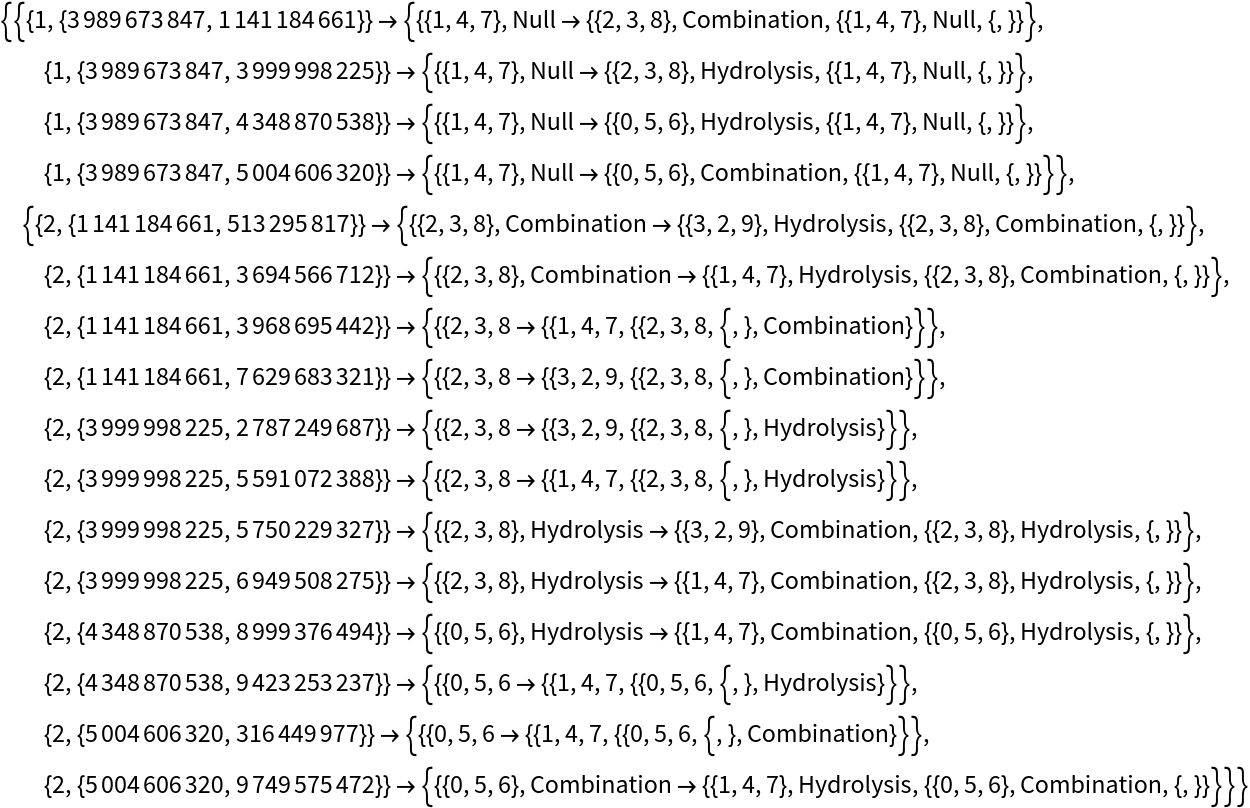 |
Vertices of a states graph can be weighted by their relative rate of occurrence at each time step:
| In[73]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, p4}, "Transitions" -> {t1, t2, t3}, "Arcs" -> {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}|>, {1, 2, 1, 2}, 3, "StatesGraph", "IncludeStateWeights" -> True, VertexLabels -> "VertexWeight", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/770d27e38202a137.png) |
| Out[73]= |  |
Vertices can also be weighted by the number of distinct evolution paths that lead to them:
| In[74]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {p1, p2, p3, p4}, "Transitions" -> {t1, t2, t3}, "Arcs" -> {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}|>, {1, 2, 1, 2}, 3, "StatesGraph", "IncludeStatePathWeights" -> True, VertexLabels -> "VertexWeight", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/51602f5f8f06ad1e.png) |
| Out[74]= | 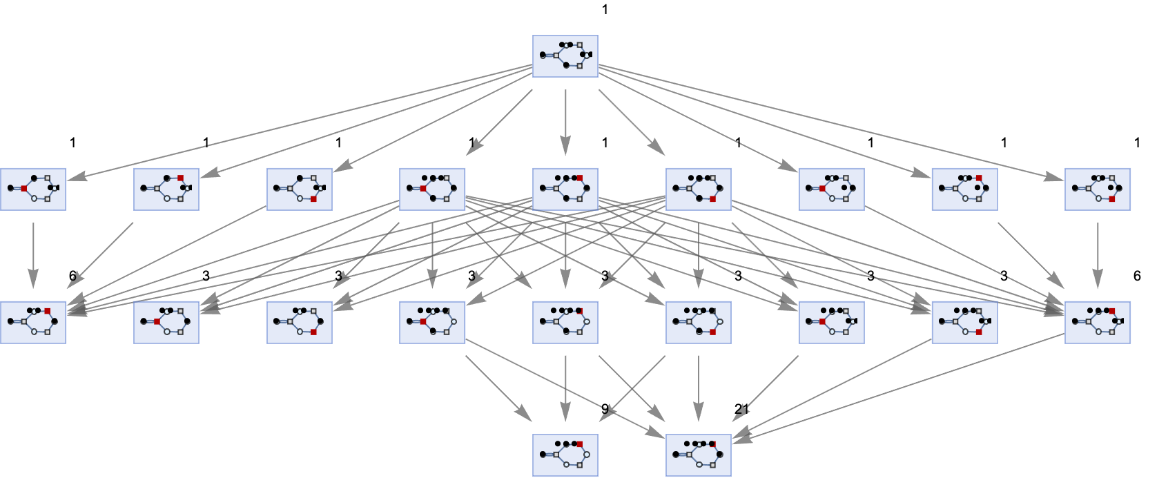 |
By default, "CausalGraphInstances" returns all possible causal graphs:
| In[75]:= | ![Graph[#, VertexSize -> 1] & /@ Take[ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {2, 0, 0}, 3, "CausalGraphInstances"], 10]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/600bd894281c354e.png) |
| Out[75]= | 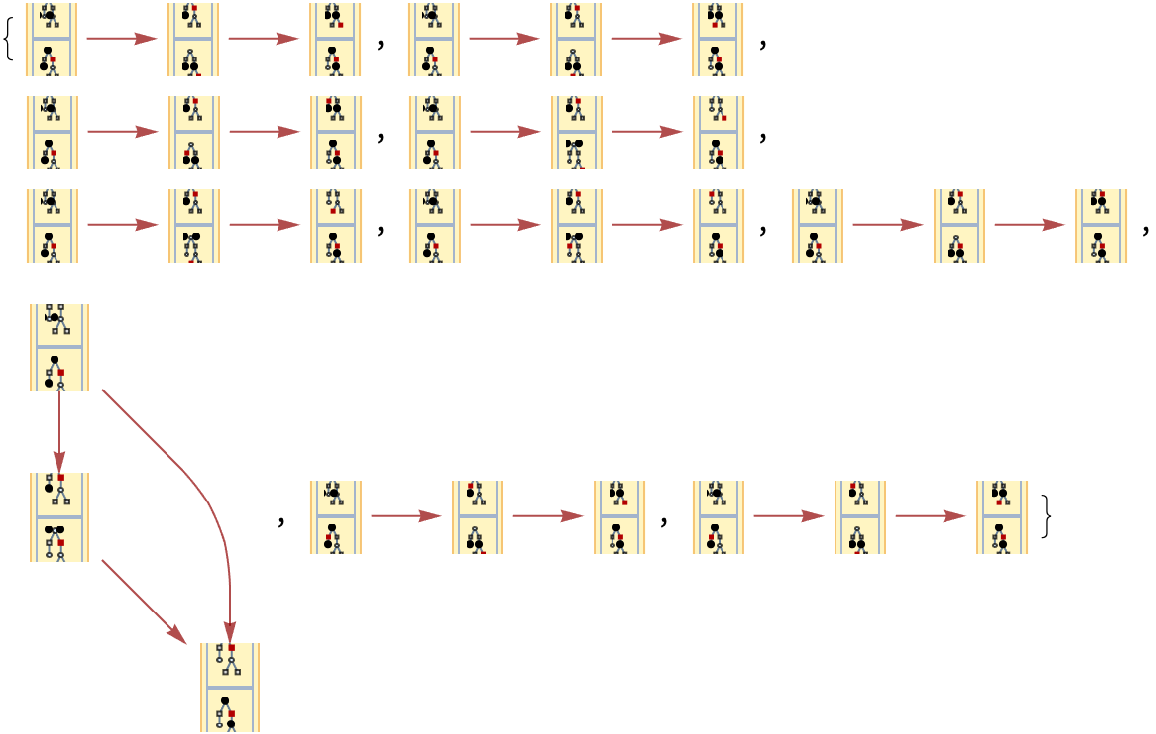 |
The number of causal graphs returned can be limited using MaxItems:
| In[76]:= | ![Graph[#, VertexSize -> 1] & /@ ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3}, {t1, t2, t3, t4}, {p1 -> t1, t1 -> p2, p2 -> t2, t2 -> p3, p3 -> t3, p3 -> t4}, {2, 0, 0}, 3, "CausalGraphInstances", MaxItems -> 5]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/7072ba570e3c4b3b.png) |
| Out[76]= | 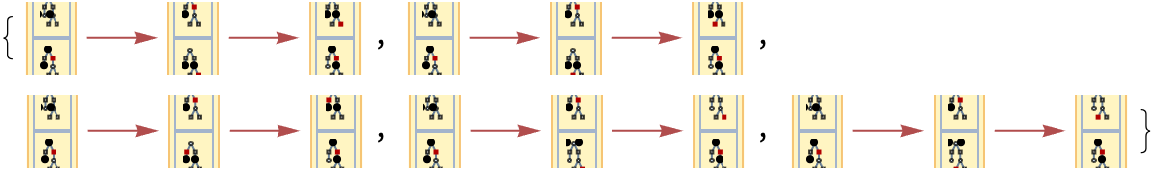 |
By default, "BranchPairsList" returns only a list of branch pairs:
| In[77]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "BranchPairsList"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/3ec68452425c02a2.png) |
| Out[77]= |  |
Common predecessor states can be shown using "GivePredecessors":
| In[78]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "BranchPairsList", "GivePredecessors" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0dc6c0a5d66d3eea.png) |
| Out[78]= |  |
Similarly, "BranchPairResolutionsList" by default lists only resolved and unresolved branch pairs:
| In[79]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "BranchPairResolutionsList"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/62d8dfeacd452a5a.png) |
| Out[79]= |  |
Common resolvents of resolved branch pairs can be shown using "GiveResolvents":
| In[80]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "BranchPairResolutionsList", "GiveResolvents" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/04a44729d9384fe8.png) |
| Out[80]= |  |
Show both common predecessors and common resolvents, where appropriate:
| In[81]:= | ![ResourceFunction[
"MultiwayPetriNet"][<|"Places" -> {"O2", "H2O", "H2"}, "Transitions" -> {"Hydrolysis", "Combination"}, "Arcs" -> {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2", "H2" -> "Combination"}|>, {1, 4, 7}, 2, "BranchPairResolutionsList", "GivePredecessors" -> True, "GiveResolvents" -> True]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/71ee24cdcb68b034.png) |
| Out[81]= |  |
By default, non-branch pair states are not shown as part of the branchial graph:
| In[82]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "BranchialGraph", VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/658d8591d83380b4.png) |
| Out[82]= | 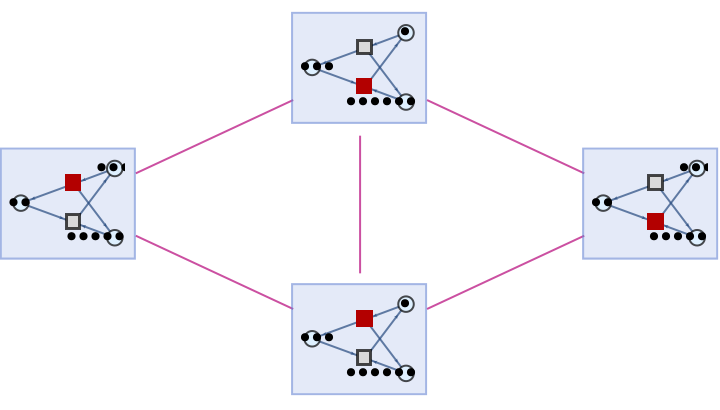 |
They can be shown using "IncludeFullBranchialSpace":
| In[83]:= | ![ResourceFunction[
"MultiwayPetriNet"][{"O2", "H2O", "H2"}, {"Hydrolysis", "Combination"}, {"Hydrolysis" -> "O2", "O2" -> "Combination", "Combination" -> "H2O", "H2O" -> "Hydrolysis", "Hydrolysis" -> "H2",
"H2" -> "Combination"}, {1, 4, 7}, 3, "BranchialGraph", "IncludeFullBranchialSpace" -> True, VertexSize -> 1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0afe1729cde5f0f6.png) |
| Out[83]= | 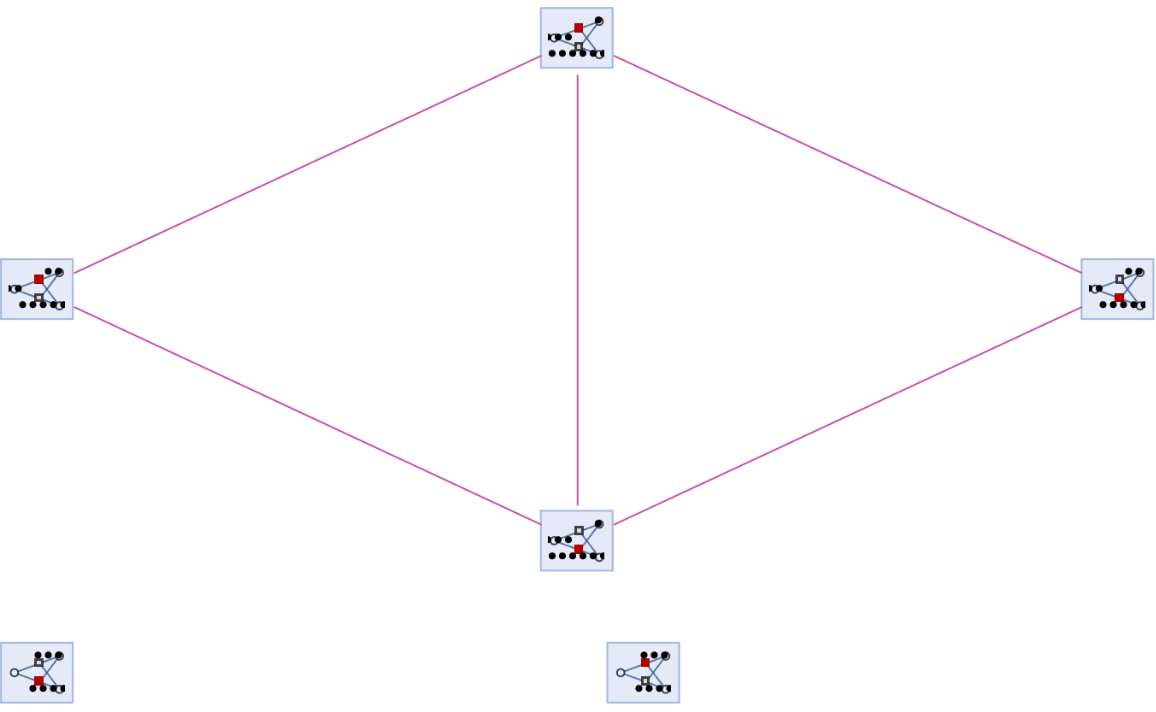 |
MultiwayPetriNet supports the same options as Graph:
| In[84]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, "LabeledGraphs"]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/3db2e85c67f7a708.png) |
| Out[84]= | 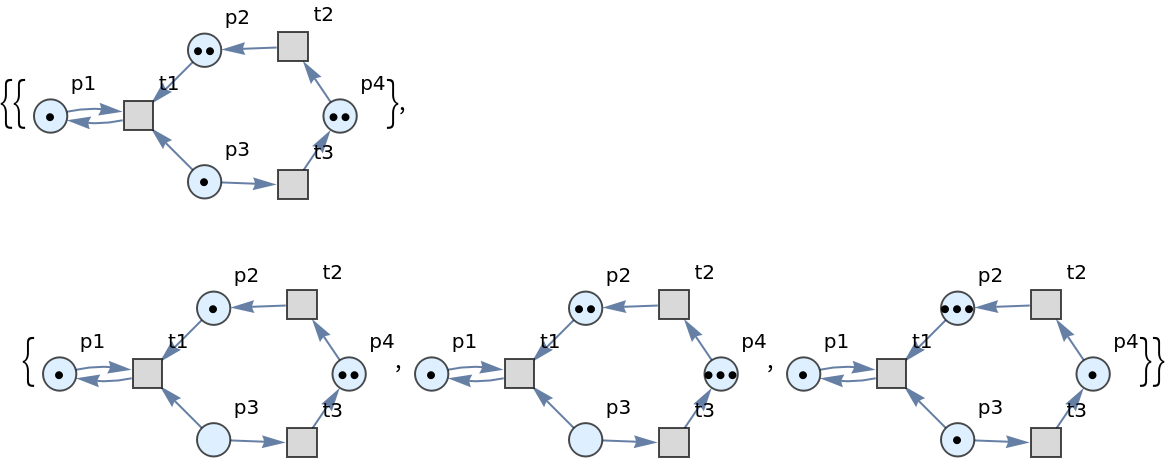 |
| In[85]:= | ![ResourceFunction[
"MultiwayPetriNet"][{p1, p2, p3, p4}, {t1, t2, t3}, {p1 -> t1, t1 -> p1, p2 -> t1, p3 -> t1, t2 -> p2, p3 -> t3, t3 -> p4, p4 -> t2}, {1, 2, 1, 2}, "LabeledGraphs", VertexSize -> 0.1]](https://www.wolframcloud.com/obj/resourcesystem/images/b55/b55a934e-81be-4579-b352-8224d36490ad/0a165cb9e5e92e1f.png) |
| Out[85]= | 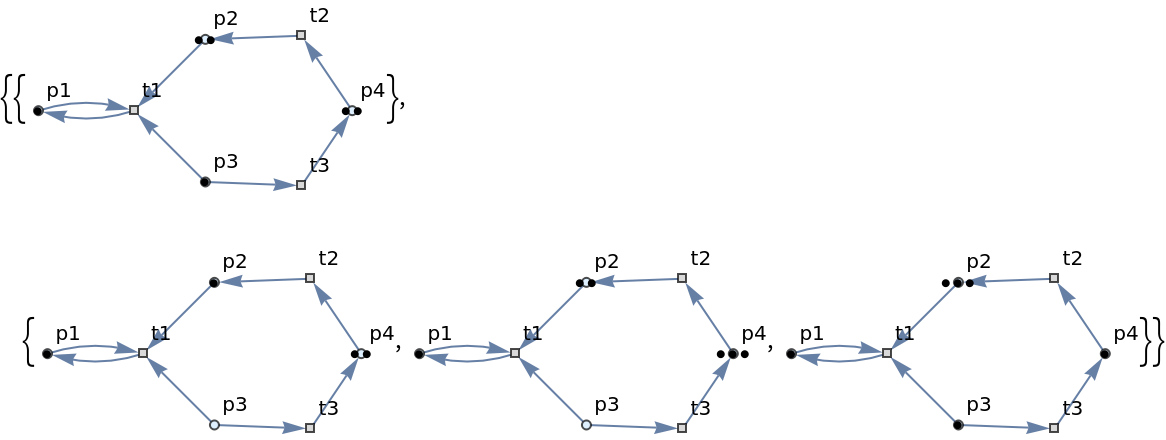 |
This work is licensed under a Creative Commons Attribution 4.0 International License