Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Measure the similarity between two molecules
ResourceFunction["MoleculeFingerprintSimilarity"][mol1,mol2] returns the fingerprint similarity between molecules mol1 and mol2. |
| "FingerprintType" | "RDKit" | the algorithm to use when encoding the molecule |
| "SimilarityMeasure" | "Tanimoto" | the bit vector similarity measure to use |
| "AtomPairs" | atoms are typed based on atomic number, number of pi electrons, and vertex degree, and all pairs of atom types, together with the distance between them, are hashed and corresponding bits in the fingerprint are set |
| "MACCSKeys" | 166 bit structural key descriptors in which each bit is associated with a SMARTS pattern |
| "MorganConnectivity" | extended-connectivity fingerprints, atoms are typed based on atomic number, heavy-atom degree, mass number and ring membership and the neighborhood around the atoms are used to set the bits |
| "MorganFeatures" | atoms are typed based on chemical features, such as H-bond acceptor/donor, aromaticity, acidity, etc. |
| "TopologicalTorsions" | similar to "AtomPairs", but rather than pairs of atoms, all sets of four consecutively bonded atoms are used to generate the bits |
| "RDKit" | identifies all subgraphs within a particular range of sizes, hashes each subgraph to generate a raw bit ID, mods that raw bit ID to fit in the assigned fingerprint size and then sets the corresponding bit |
| "Asymmetric" | (a&b)o/min(ao+bo) |
| "BraunBlanquet" | (a&b)o/max(ao+bo) |
| "Cosine" | (a&b)o / |
| "Dice" | 2(a&b)o/(ao+bo) |
| "Kulczynski" | ((a&b)o (ao+bo)) / 2aobo |
| "McConnaughey" | ((a&b)o(ao+bo)-aobo) / aobo |
| "Russel" | (a&b)o /ao |
| "Sokal" | (a&b)o/(2ao+2bo-3(a&b)o) |
| "Tanimoto" | (a&b)o/(ao+bo+(a&b)o) |
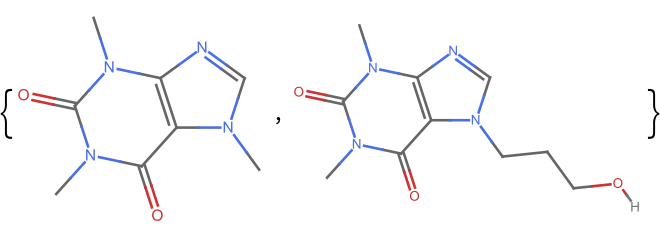
Get the fingerprint similarity between two similar molecules:
| In[1]:= |
![m1 = Molecule["caffeine", IncludeHydrogens -> False];
m2 = Molecule["7-(3-Hydroxypropyl)theophylline", IncludeHydrogens -> False];
ResourceFunction["MoleculeFingerprintSimilarity"][m1, m2]](https://www.wolframcloud.com/obj/resourcesystem/images/0af/0af7e912-f6d6-4a47-a3f5-931de7b85aab/5535298970a1a24e.png)
|
| Out[1]= |
|
| In[2]:= |
|
| Out[2]= |
|
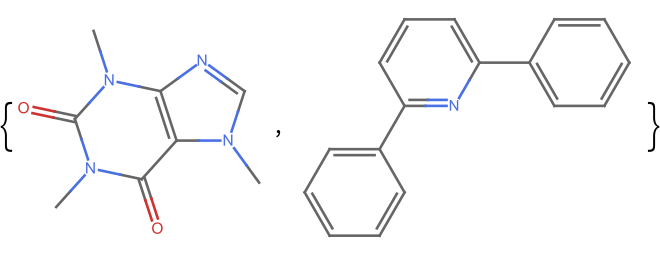
Get the fingerprint similarity between two dissimilar molecules:
| In[3]:= |
![m1 = Molecule["caffeine", IncludeHydrogens -> False]; m3 = Molecule["2,6-diphenylpyridine", IncludeHydrogens -> False];
ResourceFunction["MoleculeFingerprintSimilarity"][m1, m3]](https://www.wolframcloud.com/obj/resourcesystem/images/0af/0af7e912-f6d6-4a47-a3f5-931de7b85aab/1e9ed4ac73bf2263.png)
|
| Out[3]= |
|
| In[4]:= |
|
| Out[4]= |
|
MoleculeFingerprintSimilarity works on molecules created from any source, from MoleculeRecognize to Entity:
| In[5]:= |
![ResourceFunction["MoleculeFingerprintSimilarity"][
MoleculeRecognize[\!\(\*
GraphicsBox[
TagBox[RasterBox[CompressedData["
1:eJzt3XtwVGWax/G47h9eAbG8lH8IlqUlNbNAydRqleUCRdUOYxHJCkihKHjJ
UCwqKKMUM8yCsJY1JQILRdXIHXGQQZSblwGj4TqDghuILCEZQyMZIZFLBwYm
hASf7bcx5JxOn9PnJH366T79/VS9Isl7ut9Ok1+//Zz3PX3H0+Mf+eU/FRQU
TLwq9p9HnprU/6WXnpo8pEvsL8NemDh2zAvFz/7ihV8Vjyl+6f6nr4x9sfsV
BQVPx9o/x/5fAAAAAAAAAAAAAAAAAAAAkJ8aoiL7vhDZuEFk86ZY2xr7e6VI
tEF7ZAAQbg3HRN58WsQs6Xdqw2aKHIxqjxQAwqdynXv+JrY3tmmPGADCo3x5
25xduEGktjY2R26I/RkR2fx22z4LyrVHDgAhUGHP1p/NFHEsO8QyeVGRvf/B
85kcLACEz5rRllx909sxr1hyuM+iYMcHAKEWsc9t99Z7O+x8mf24SJBjBIAQ
+9Zybq7PfH/H/te/tB67uiqY8QFA2FWtas3Soe/5O3b3/7Qe+5vtwYwPAMKu
/Pftn9NaM3yQzwwHAFyy/XeWOe1uf8d2ZC4NALjk08npmQ8XvhPM+AAg7KxZ
+uxGf8da68Nv+JxLAwAuqd9iWX/2W3/HWtcQb/g2mPEBQOidEilqR57a8jvW
jgU7SgAIte2WGnFBoYdMjWX38ALO0QFA2iTMiQvuFdlbm7xrtCyhL3NhAEiL
aMI+ZdMenSKydZ9I5UGRfVtF5j/Vts8m6sIAkDa1u/xdf3hTpfaIASCctv1R
pKi3Q/4WiqzYFr/8JQAgYOYz6g4fFolGL10Pns+mAwAAAAAAAAAAAAAAAAAA
AAAAAAAAAAAAyFnTpk2T3r17y+jRoyUSiaS9PwDAmcnRgoKCy2327Nlp7Q8A
cFdaWmrL1b59+6a1PwDAHTkMALrIYQDQRQ4DgC5yGAB0kcMAoIscBgBd5DAA
6CKHAUAXOQwAushhANBFDgOALnIYAHSRwwCgixwGAF3kMADoIocBQBc5DAC6
yGEA0EUOA4AuchgAdJHDAKCLHAYAXeQwAOgihwFAFzkMALrIYQDQRQ4DgC5y
GAB0kcMAoIscBgBd5DAA6CKHAUAXOQwAushhANBFDgOALnIYAHSRwwCgixwG
AF3kMADoIocBQBc5DAC6yGEA0EUOA4AuchgAdJHDyFmNzXLmVFOsNct5Twe0
9G/y2D8dWu/zzNmLHvpfbO3f6KU/woAcRq6qfKtReg5sire3y39I2T+ytFF+
+vOmeFvooX86WO/TtL+l6N9Y0dr/3+Y1Z2SM0EcOI1dZM85vDnvpnw6JOTxk
lnu2WnP4sQXkcL4gh5GrcjGHTfujy32Tw/mJHEauytUcNq3aoT85nJ/IYeSq
XMzh/oN+rP061CfI4fxEDiNXZSyHzzbLdzVNl5u3dQ9t73PslAsp6xPkcH4i
h5GrrBm3qsJD/9X+cvh8XaMsnnQhaV1hyJRmqT7lL/vXHmqWndPtt5dYnyCH
8xM5jFxlzTiTi598fEE+WefQYt/77xFNnnP47+XJ67qJbV2F++3Y5+DmKxfk
P/+99fi+M+xza3I4P5HDyFWJtdeeLi0xP11zuNGelT+N5ff28mY5c7ZJDsXy
edKghHNujd7G2HKff9/T6DgWcjg/OeXq0aNHpUePHvGvderUSdavX+/aH8g0
p7UIXppbDh/9qPV2f1Icy982PWJza0sWL9zjXC9OXpO+KKVTktcnyOH8lJir
Dz74oEyfPl2uuuoq29dNGz16tLz77rvkMLKCNeOmlzTL6ZNN8n1tc9Jm5rKl
ltqscw5ftNVw1x5K3stat3DLS8dzg2cbk9YnOprD0WhUli1bJpFIxPex0FNW
VmbL1WuvvbZN/lrb9ddfb/v7+PHjtR8C8pT9PF261kvEcnjeBRlZ7F5zsObl
I+3JYUlWn4h9sbr9OWwyuHv37vHfyy5dusjSpUt9HQ9dDz/8sFx55ZWu+ZvY
rrjiivhzzusutOitH7bPmc1cvH332bY+sdvjPDuZUaNGtfk9nTBhQjseHzLp
5MmTMmbMGMesveGGG+S5556TXr16Ofbp169ffE4NZJpWDlvnwqZ9XON8Wynv
s9Fen7A2Pzmc+L7W2nr37h2fKyP7zJkzJ/7exem5Gzt2bDynW5j3OJ07d3bs
b2rHPNfIJJUcrmuUYT9vW9ftyH1erk8Utj+HzXzI7f2r+V0354KQHUpKSuSu
u+5yfL4eeOAB2b9/f9JjTc5OnTrV9bmeNm1ahh8R8lXGc/hUo4xMmLO6rVnz
fp/2+kR/nzm8du1az/VEM/+CnurqaiksLHR8fkytd82aNZ5uy9SEzToJt9sy
/zaAIGUyh88fudAmgyvOpnGMiWuWfeRwy7k5r62oqIj3rhl29uxZefnllx2f
E7M+YsaMGe26bfM+p1u3bq61Y87jISiZyuGjf2m7TtlLBvu9z8T1E15y2O39
qVszNWPO62TGkiVL5JZbbnF8Lh577LH4fo2Omj17tmvt2Jyz5fUX6Wa9XkSq
/cXx/uta+7tdA7jVRSlP3Csyoln+lqIW4XSfqdfW2ddhjFmaovYcm+O4neNJ
1VjbFqxdu3a5rnEw3zN90snkrFlL7PacU5tCuv3ww6WWfhdlz1sJn6UxI9ne
Oj3J1qm1p5lz7EgfM7d9/PHHHX/eZm68ePHiQMdg3uukqh1z3hbZrnK1PYPH
LM2uPcaJ+2A72kydghpix5kar9teOFMjNrXiTDHn6dxqx+ZcAc87slFjtT2D
3fZpaEm1Tq29dQrmSO1j1ji4nS8dNGhQfK2EFnMewa12bNa5UTtG9miWDePs
e9yGTGmSKZMuyCvJ2rgmWbQ1szltarrpzuDE30l4Y9b4mrW+Tj9Ls0bYrBXO
BiZn3WpZ5nXYXJsEUHe27RrhVM3t+hLpZr2GRJCNtW3uzB43s9fNLdPM+oVs
lKp2bGpUvC+Cqnbk8JjV3j8nqaPau06tPc3kPWvb2po7d278mg9OPzdzrQjr
XuRsZd5XudWOzflbaseAXUfXqbWnsbat1ZYtW+See+5x/Fm57UXOVi17pJ1q
xy17pHlvBFziti406JbPa9vM+TVzTUqnn42ZU65evVp7mB1iXuPdasfmvRG1
Y0Bca3qZaPm2ts2sL5s0aZLjz+Oaa66RV199VRoaGrSHmjamLsz1NQFnbte2
zGSdIh/O4Zi536233ur4cxgxYoTU1NRoDzMwXF8TcGbmo6Y+4fY7kokW1rVt
e/bskT59+jg+7iD2Imcrrq8JpGbmLJq1ijCtbTN7kZ944gnHx3rzzTfLokWL
tIepwrz2Dx482PFnw/U1Ad05chjWtr322muue5EnTpwoZ85k05VFdHB9TcAb
jTlyrq5t++CDD+SOO+5wfFwPPfSQVFVVaQ8z63B9TcAbjTlyrpy7qaiocH2t
yqa9yNmK62sC/mRyjpzNa9vMHrdx48Y5jt28Zs2aNUt7mDnFy2cz5cP6GsAr
U8c1a/WDniObuVC2nbeZN2+edO3a1XHMxcXFcvz4ce1h5iyurwn4Y95Tmjmy
23r9dLRsWNPkZS/y3r17tYcZGlxfE/Av6DmyOYeu8Xt3+PDh+BzMaVy33367
rFq1KuPjygdcXxNonyDnyJlc22b2Ik+ePNlxLFdffXV8zhamvcjZKtX1NbP1
uqBANghqjhz02rbly5fLbbfd5nj/w4cPD/Ve5GzldH1N85rvX+z1s3KvyOZN
re3LfSK17XzPZd6rmeb1ZbnBZ3+gg4KYIwexts3LXuQdO3ak9T7hT7Lra5p1
b97Fgu+9uRI70Ln961CRrZU+bvJg67H3rvFwQIWl/zs+xg6kR8scOR1ZnK61
bXV1dfLkk0863s9NN90kCxYs6PiDR9qY593ksWmeRWN5eb9L/ia2wvke56uW
XB36XgD9gWCYeY2p67mtUfLSOrq27fXXX5frrrvO8fZffPFF9iKHQbSsbc7+
aqFI5eFY1sbCNnpM5Ms/iQxPzGIv1wKx5OognznsqT8QPLM+v6NzZL9r29at
Wyd33nmn4+0NGDCAvcih0ZCQr4Uie+udu297y57Fb6f6d0AOIzw6Okf2srYt
1V7ku+++Wz788MMMPWJkRNUqS67eK3LMwzEbJ9hz2/WfFTmMcGrvHNlpbVt9
fb08//zzjsd16tRJZs6cqfBIEbgxlrntvHKPB50SKbIct+Fbl77kMMKtvXNk
69q2+fPny4033ujY95lnnmEvcmhFfMxrE2z/ncfzaX7Pu0XIYeQsv3PkgQMH
So8ePRy/f99997EXOfQsGVlQ7O/Q+i2WY3/r8T4KL61F3rgheTPfe+/X5DBy
Xssa0vbWkc3nxa1cuVL7YSATzpf5nKtaWfN1mMsaNms/n40cRgiYNWtun7WT
2KZMmSLnzp3THjYypc46p33T58HkMOBHy7p+pznysGHD2Iucj+o+yex8+Gcz
W/c4O7XDW8hhhJ75PB1rBvfv3197SFBjzdKX/R1av6v12D7zvd2H3/105DBC
asWKFbYcHjlypPaQoOaYfS9zxMehn1rWED+70aUj69aAROQwbBYN9JinVsfs
ddytdS59yWEgETkMG9u5ulSZ+qNFRfb1bq7X+/GbqwfIYYQeOYw2/jDYnsWr
9zl0PCXy5i/sfTelym1yGEhEDqOthH3KLWsgNmwTqTx46Zrwya5LPGWzh9u2
5HAhOQwY5DCSOybyy17e1/a+4SWDjQP+cvgf1IcRfuQwXFVuEXnlIef8nbla
pNbPBxZZcrjYy3lAS/9h5DDCiRyGN7Gsra299Jl05k+FzwMHwoocBgBd5DAA
6CKHAUAXOQwAushhANBFDgOALnIYAHSRwwCgixwGAF3kMADoIocBQBc5DAC6
yGEA0EUOA4AuchgAdJHDAKCLHAYAXeQwAOgihwFAFzkMALrIYQDQRQ4DgC5y
GAB0kcMAoIscBgBd5DAA6CKHAUAXOQwAushhANBFDgOALnIYAHSRwwCgixwG
AF3kMADoIocBQBc5DAC6yGEA0EUOA4AuchgAdJHDAKCLHAYAXeQwAOgihwFA
FzkMALrIYQDQRQ4DgC5yGAB0kcMAoIscBgBd5DAA6CKHAUAXOQwAushhANBF
DgOALnIYAHSRwwCgixwGAF3kMADoIocBQBc5DAC6yGEA0EUOA4AuchgAdJHD
AKCLHAYAXeQwAOgihwFAFzkMALpmzZply+FHH31Ue0gAkBei0ahMmDDBlsGm
9ezZU3toABB6c+bMkS5durTJYNMGDBigPTwACK3S0lLp3r170vw1rWvXrlJT
U6M9TAAInUgkIv369XPM327dusnatWu1hwkAoeNUA25pnTt3lqlTp2oPEwBC
admyZY41YNNGjRoVz2kAQHqZGnDv3r0d87dv375SVlamPUwACB1TAy4qKqIG
DAAZZmoL06ZNS1kDpgYBAOnnpQZs5skAgPTyUgM2fQAA6WXmtqNHj3atAS9d
ulR7mAAQOi01YKcaBDVgAAiOqQG77UUePHgwNWAACICp77rtRe7Vqxc1YAAI
QKoasKlBUAMGgPRLVQM2jRowAATD7HOjBgwAmWeu80ANGAAyz9QWqAEDgI5U
NeDx48dTAwaAAKSqAZu9yNSAASD9UtWAzV5kasAAkH5ePpNo9uzZ2sMEgFBy
+1x6asAAEJxUn0tPDRgAgsHn0gOADj6XHgD0pKoB87n0ABAMsw4tVQ2Yz6UH
kDOaRE5E6+Pt9Hlvh0R/7B895++uThw5In8t2yGV2z+TA9t3SmXZ13Lku3r5
h4/bMHVgpzkwNWAAuehM1R/ky4UFl9pH36TuX73mcv8vNuz3dB+R7Utld8t9
JG1Dpax0v6c8NushqAEDCBNrrvrNYS/9q9b/h0v+JrRVn8tpD2M210CjBgwg
LILM4eN/mWPP2RVz5WD5X+VYbb2cqK2TQ+Wfyd7lCVn8p9RjMLUJsw+OdcAA
wiDIHP7m/fsv993zkXMN45sPR7Te5oLlctz3owCA3BVcDjfI/pa57oLnJOJ6
DrBWyiy14m98nv8DgFwWaA4vaa037D/kfrtV6yfLVyunS9matXLU47oNAAgD
W656qM1eOOyjLmE7RzdU9m3/Wk6QsQBgY8vhxXPlwPbPpaL046TNfG//+494
zuHTkZKk6yJ2r5wrX5fulOpInec1ywAQVrYc9ts81DFOHng/5e2YXD5QXuNr
PwcAhEXQORx3rlaqSlLt5TDn8+ZKhHN0APKMLYdX/Vm+P1Mv39c6tGiD1FjX
BHvNYYvT0To5VLZDyte9mjyXF7wlR5sCeKAAkKX8nqdr8Lm+IpUT31XJ/1lr
zrH2vzu/7/DtAkCuCGrd2umImfP+XspW/FoOfpt6HJHNky/f7hdrv/LzEAAg
pwWVw5FPWtes7fm0JuXtXqj5yLJuY5VwxQgA+SKoHK7/eoml5js9xX66hP4e
r+MGAGEQ2H6681XyVcJaiOra5F2jkRLbObuynV6uugYA4RDkdX5qE6+3ZtYK
r5gu+0o+l8o/75SKknel7J0hCesllktduh4cAOSAoK8/HPlsmvf1yAt+I9Vn
0vGoACB3WHN1z6ce1q0dtpxP87DOzYhGdkv56nEuGfy4lJXslBOsGwaAYMVy
NlpbJ8ciR+LXgj8WqYvvDWnUHhcAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
AAAAAAAAAAAAAAAAAAAAAADS4v8BOPMFxg==
"], {{0, 144.}, {177., 0}}, {0, 255},
ColorFunction->RGBColor,
ImageResolution->144],
BoxForm`ImageTag["Byte", ColorSpace -> "RGB", Interleaving -> True],
Selectable->False],
DefaultBaseStyle->"ImageGraphics",
ImageSize->Automatic,
ImageSizeRaw->{177., 144.},
PlotRange->{{0, 177.}, {0, 144.}}]\)], Molecule[Entity["Chemical", "LCysteine"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/0af/0af7e912-f6d6-4a47-a3f5-931de7b85aab/3b163bdd349a05ee.png)
|
| Out[5]= |
|
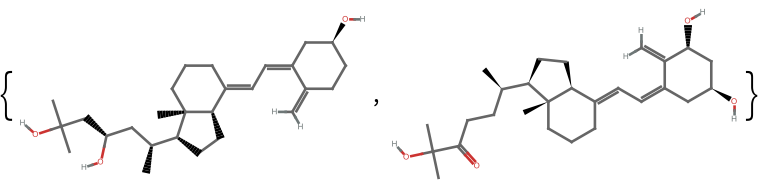
The fingerprint method and similarity measure used can greatly affect the calculated similarity. Take two nominally similar molecules:
| In[6]:= |
![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/34dd38ec-ee23-489e-b650-277501df23ef"]](https://www.wolframcloud.com/obj/resourcesystem/images/0af/0af7e912-f6d6-4a47-a3f5-931de7b85aab/0b2c85d014169a64.png)
|
| Out[6]= |
|
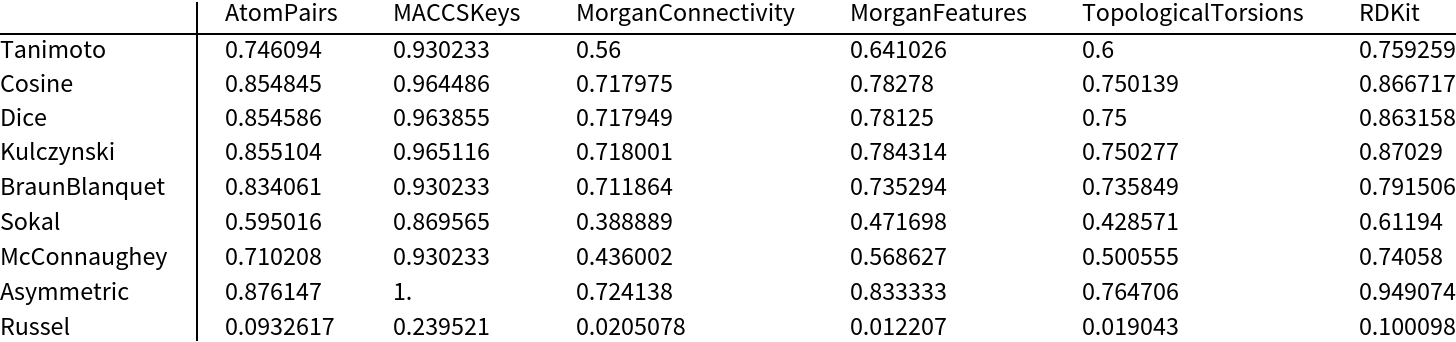
Measure the similarity using all available fingerprint types and similarity measures:
| In[7]:= |
![data = Table[
ResourceFunction["MoleculeFingerprintSimilarity"][m1, m2, "FingerprintType" -> fp, "SimilarityMeasure" -> sim], {sim, smeasures = {"Tanimoto", "Cosine", "Dice", "Kulczynski", "BraunBlanquet", "Sokal", "McConnaughey", "Asymmetric", "Russel"}}, {fp, fptypes = {"AtomPairs", "MACCSKeys", "MorganConnectivity", "MorganFeatures", "TopologicalTorsions", "RDKit"}}];
MinMax[Flatten[data]]](https://www.wolframcloud.com/obj/resourcesystem/images/0af/0af7e912-f6d6-4a47-a3f5-931de7b85aab/1b62a4b7461cd94b.png)
|
| Out[7]= |
|
Visualize the results in a table:
| In[8]:= |
|
| Out[8]= |
|
MoleculeFingerprintSimilarity returns 0 for completely dissimilar molecules:
| In[9]:= |
|
| Out[9]= |
|
MoleculeFingerprintSimilarity returns 1 as the result if the two given molecules are identical:
| In[10]:= |
|
| Out[10]= |
|
The presence or absence of explicit hydrogens in the molecular graph can influence the computed similarity:
| In[11]:= |
|
| Out[11]= |
|
| In[12]:= |
|
| Out[12]= |
|
Create molecules from the list of central nervous system (CNS) agents obtained from PubChem:
| In[13]:= |
|
Find the five nearest molecules to the tranquilizer diazepam:
| In[14]:= |
|
| Out[14]= |
|
| In[15]:= |
![Nearest[CNSagents, diazepam, 5, DistanceFunction -> (1 - ResourceFunction["MoleculeFingerprintSimilarity"][##] &)]](https://www.wolframcloud.com/obj/resourcesystem/images/0af/0af7e912-f6d6-4a47-a3f5-931de7b85aab/15f9ed8e90592a65.png)
|
| Out[15]= |

|
This work is licensed under a Creative Commons Attribution 4.0 International License