Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Compute the molecular complexity of a given molecule
ResourceFunction["MolecularComplexity"][mol] returns the molecular complexity Cm for Molecule mol. | |
ResourceFunction["MolecularComplexity"][str] returns the molecular complexity for the input molecule identifier str. |
A molecule identifier readable by Molecule or a Molecule object itself can be taken as input. By default, it returns a numerical output corresponding to the Cm of the molecule:
| In[1]:= |
| Out[1]= |
Use a Molecule as input:
| In[2]:= |
| Out[2]= |
| In[3]:= |
| Out[3]= |
MolecularComplexity accepts a SMILES or InChI string argument:
| In[4]:= |
| Out[4]= |
| In[5]:= |
| Out[5]= |
MolecularComplexity threads over lists:
| In[6]:= |
| Out[6]= |
Zero complexity inputs return an error:
| In[10]:= |
| Out[10]= |
| In[11]:= |
| Out[11]= |
MolecularComplexity utilizes Molecule options, so it can only take identifiers that Molecule can parse:
| In[12]:= |
| Out[12]= |
Show that the trend in molecular complexity of alkanes with increasing (CH2) groups from 1 to 10 is linear:
| In[13]:= | ![complexities = ResourceFunction[
"MolecularComplexity"] /@ {"propane", "butane", "pentane", "hexane", "heptane", "octane", "nonane", "decane", "undecane", "dodecane"};](https://www.wolframcloud.com/obj/resourcesystem/images/d40/d40d19dc-e238-4a94-9b06-eee5a0cc25a8/2a51211fc6319bfc.png) |
| In[14]:= | ![Show[ListPlot[Thread[{Range[1, 10], complexities}], AxesLabel -> {"\!\(\*SubscriptBox[\(CH\), \(2\)]\) groups (n)", "\!\(\*SubscriptBox[\(C\), \(m\)]\)/mcbit"}, PlotStyle -> Black], Plot[Evaluate[
Fit[Thread[{Range[1, 10], complexities}], {1, x}, x]], {x, 1, 10}, PlotStyle -> {Red, Dashed}], ImageSize -> Large]](https://www.wolframcloud.com/obj/resourcesystem/images/d40/d40d19dc-e238-4a94-9b06-eee5a0cc25a8/23da714338a65365.png) |
| Out[14]= | 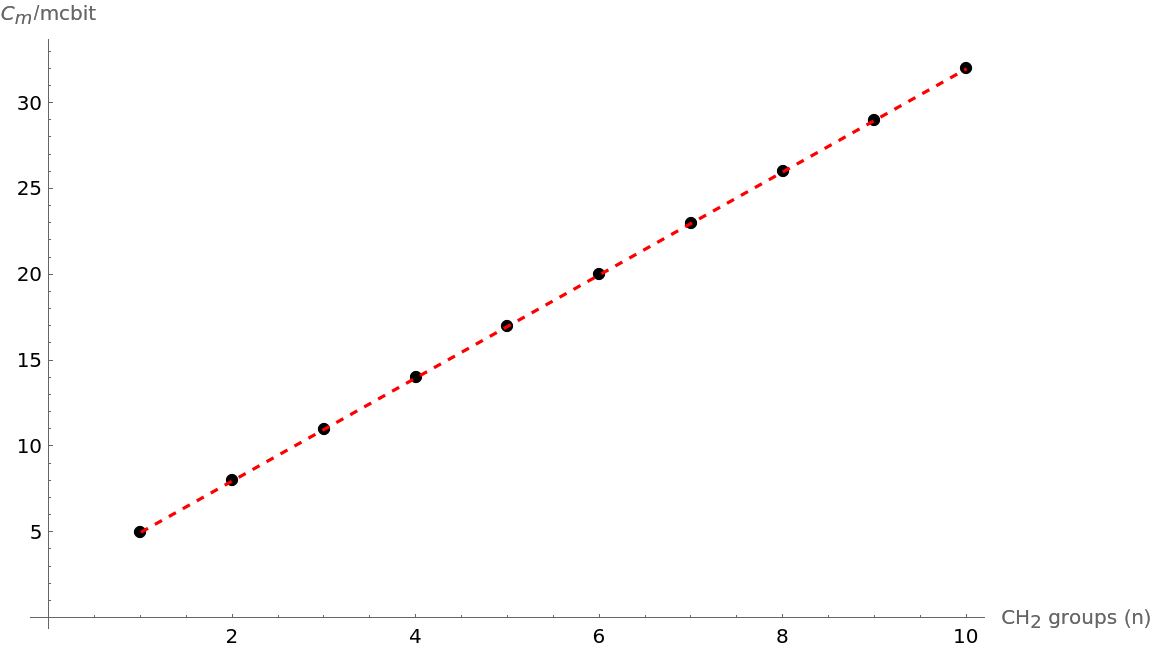 |
Calculate the change in complexity for a Diels–Alder reaction:
| In[15]:= | ![(* define the reactants and product *)
butadiene = Molecule["1,3-butadiene"];
ethene = Molecule["ethene"];
cyclohexene = Molecule["cyclohexene"];](https://www.wolframcloud.com/obj/resourcesystem/images/d40/d40d19dc-e238-4a94-9b06-eee5a0cc25a8/7d6351bf3cf15a7d.png) |
| In[16]:= | ![(* compute the complexities and find the difference *)
cButadiene = ResourceFunction["MolecularComplexity"][butadiene];
cEthene = ResourceFunction["MolecularComplexity"][ethene];
cCyclohexene = ResourceFunction["MolecularComplexity"][cyclohexene];](https://www.wolframcloud.com/obj/resourcesystem/images/d40/d40d19dc-e238-4a94-9b06-eee5a0cc25a8/0b73ce083c92681f.png) |
| In[17]:= |
| In[18]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/56784de3-8455-442e-997c-6dcfd8368fff"]](https://www.wolframcloud.com/obj/resourcesystem/images/d40/d40d19dc-e238-4a94-9b06-eee5a0cc25a8/066481d65b82a774.png) |
| Out[18]= | 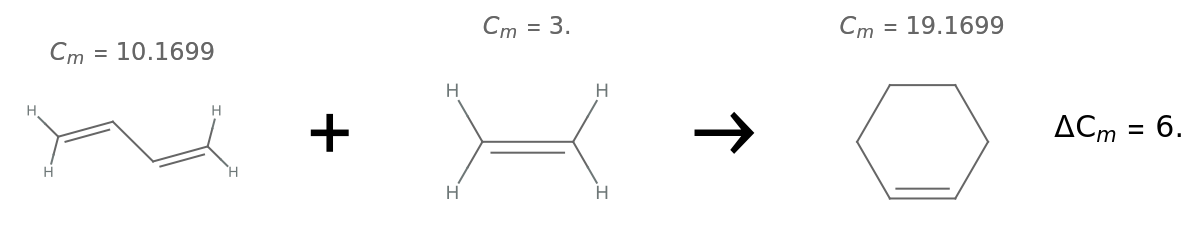 |
This work is licensed under a Creative Commons Attribution 4.0 International License