Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get information about a KEGG module
ResourceFunction["KEGGModule"]["Modules"] gives a Dataset with basic information for all the KEGG modules. | |
ResourceFunction["KEGGModule"][keggcode,prop] gives a property prop for a specific keggcode. | |
ResourceFunction["KEGGModule"][keyword,"Query"] gives a dataset for a specific keyword referring to a module. |
| "Module" | information for keggcode referring to KEGG module (i.e., M00001) |
| "Entry" | ExternalIdentifier of the module KEGG code |
| "EntryType" | the module type |
| "Name" | the module name |
| "FullName" | the module full name |
| "ReactionInName" | the reaction specified in the module name |
| "Definition" | definition of module where the comma indicates alternatives, plus represents a complex, and minus denotes a non-essential component in the complex |
| "DiagramLink" | thumbnail of the corresponding MODULE diagram link |
| "DiagramImage" | thumbnail of the corresponding MODULE diagram image |
| "Orthology" | the list of KEGG ORTHOLOGY identifiers |
| "Taxonomy" | link to the taxonomy information |
| "OrthologTable" | link to the ortholog table |
| "Class" | classification of modules |
| "PathwayExternalID" | ExternalIdentifier corresonding to the KEGG pathway map |
| "PathwayHyperlink" | Hyperlink corresonding to the KEGG pathway map |
| "Reaction" | the list of reactions (R numbers) in the metabolic pathway module |
| "Compound" | the list of compounds (C numbers) in the metabolic pathway module |
| "RModule" | corresponding pathway modules (Reaction module only) |
| "Comment" | free text comment |
| "Reference" | reference information |
Get the list of all the modules in the KEGG module database:
| In[1]:= |
| Out[1]= |  |
Get the information from KEGG about a specific module:
| In[2]:= |
| Out[2]= | 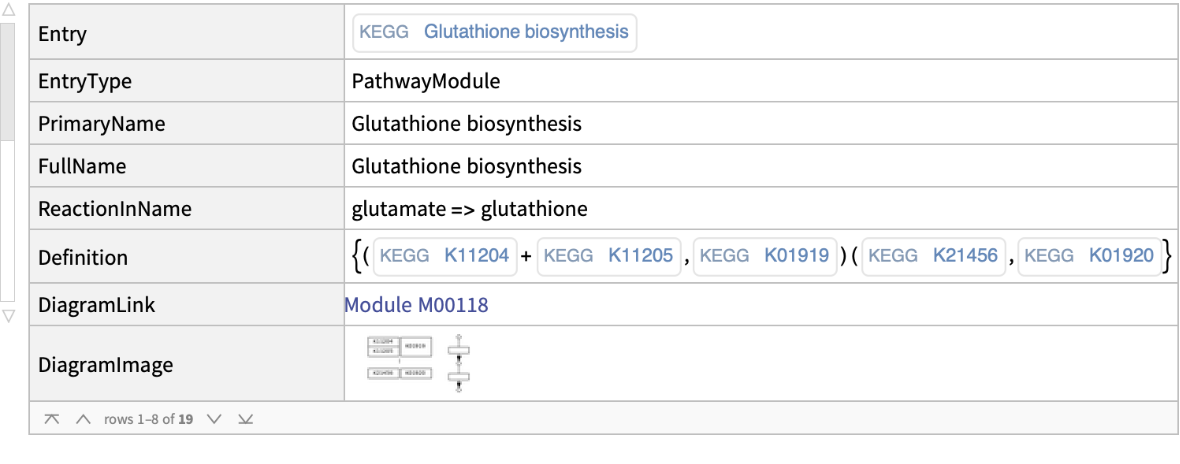 |
Get the information from KEGG about a list of modules:
| In[3]:= |
| Out[3]= | 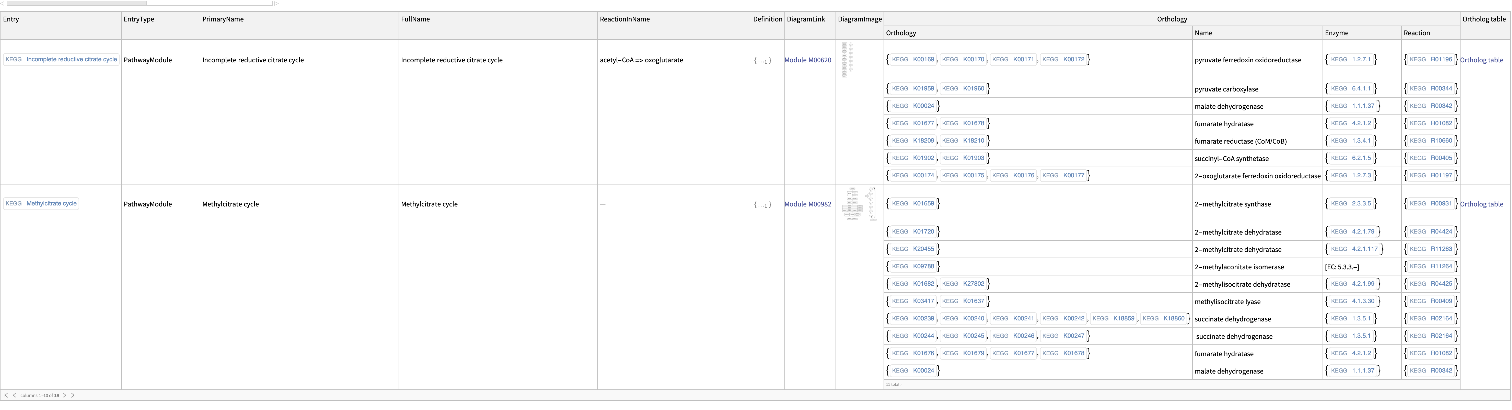 |
Get the KEGG module codes for a specific query:
| In[4]:= |
| Out[4]= | 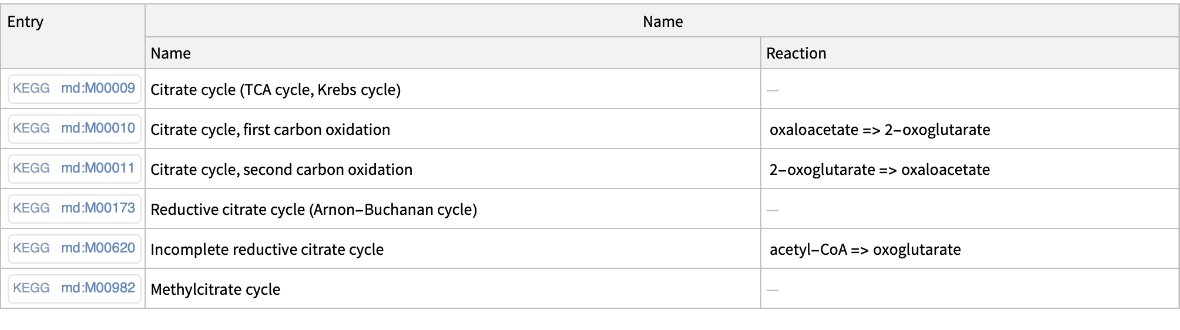 |
Keywords may have more than one word:
| In[5]:= |
| Out[5]= | 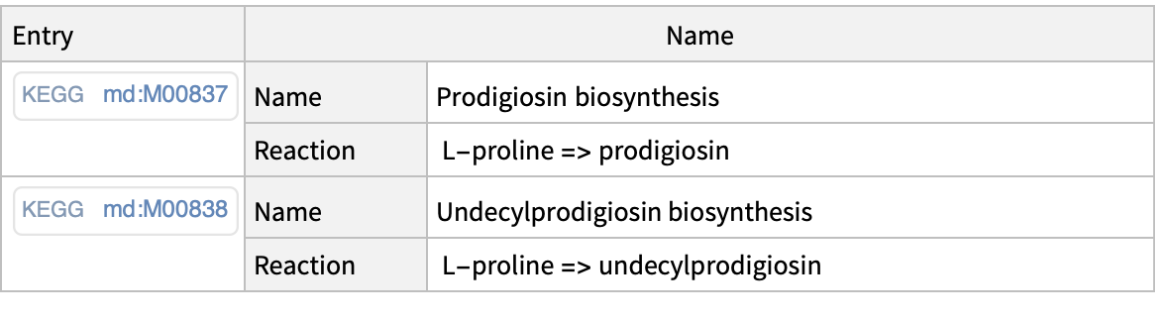 |
Get the dataset properties independently for each module:
| In[6]:= |
| Out[6]= |
Get the dataset properties independently for a list of modules:
| In[7]:= |
| Out[7]= |  |
Get information for a specific identifier:
| In[8]:= |
| Out[8]= | 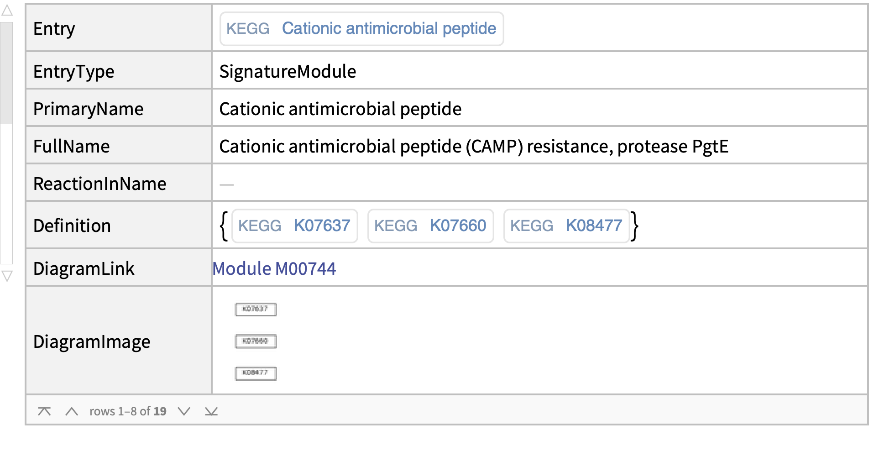 |
This is equivalent to explicitly requesting the "Module" information:
| In[9]:= |
| Out[9]= | 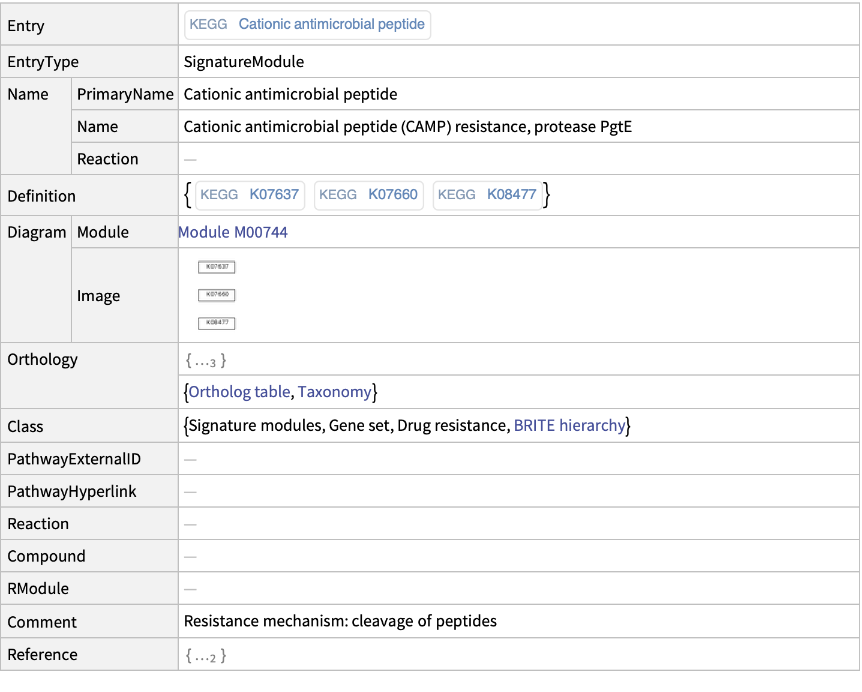 |
Only keywords known by KEGG are supported:
| In[10]:= |
| Out[10]= |
Get the pathway associated to the glycolysis module:
| In[11]:= |
| Out[11]= |  |
Get reactions associated to the glycolysis module:
| In[12]:= |
| Out[12]= |  |
Get the compounds associated to the glycolysis module:
| In[13]:= |
| Out[13]= |  |
Get the vertices and edges list associated to the pentose phosphate pathway associated with the glycolysis module:
| In[14]:= | ![vNamesRules = Normal[ResourceFunction[
ResourceObject[<|"Name" -> "KEGGPathway", "ShortName" -> "KEGGPathway", "UUID" -> "c26fb78f-9391-4e5b-8348-1b3a9bcb8293", "ResourceType" -> "Function", "Version" -> "1.2.1", "Description" -> "Get the graph and additional information of a KEGG pathway", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$cf8afda6cda14c308cd1e656f8629ece`KEGGPathway", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/81066f03-423e-4566-800b-da9d03968a36"]|>, {ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"}]]["hsa", "00030", "Entries"][
All, #EntryID -> #EntryGraphName &]];
graph = ResourceFunction[
ResourceObject[<|"Name" -> "KEGGPathway", "ShortName" -> "KEGGPathway", "UUID" -> "c26fb78f-9391-4e5b-8348-1b3a9bcb8293", "ResourceType" -> "Function", "Version" -> "1.2.1", "Description" -> "Get the graph and additional information of a KEGG pathway", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$cf8afda6cda14c308cd1e656f8629ece`KEGGPathway", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/81066f03-423e-4566-800b-da9d03968a36"]|>, {ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"}]]["hsa", "00030", "Graph"];](https://www.wolframcloud.com/obj/resourcesystem/images/beb/bebabad2-b1d9-4b1c-805e-bc3c54c4417a/4f5b9012277d6a65.png) |
| In[15]:= |
| Out[15]= |  |
| In[16]:= |
| Out[16]= | 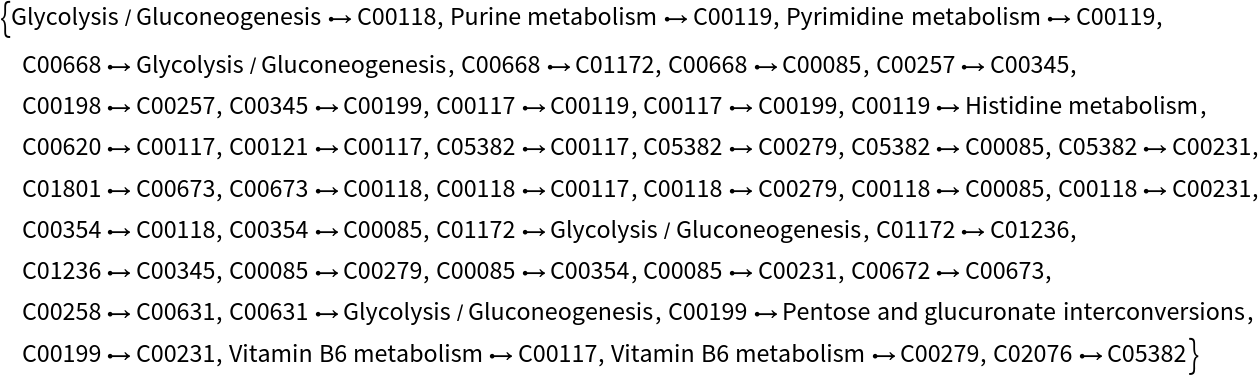 |
Using the information about reactions and compounds from the glycolysis module, we get the next list of vertices and edges to highlight in the pentose phosphate pathway:
| In[17]:= |
| Out[17]= |
| In[18]:= |
| Out[18]= |
Highlight in red (i.e.,vertices and edges) the M00008 module in the pentose phosphate pathway:
| In[19]:= | ![ResourceFunction[
ResourceObject[<|"Name" -> "KEGGPathway", "ShortName" -> "KEGGPathway", "UUID" -> "c26fb78f-9391-4e5b-8348-1b3a9bcb8293", "ResourceType" -> "Function", "Version" -> "1.2.1", "Description" -> "Get the graph and additional information of a KEGG pathway", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$cf8afda6cda14c308cd1e656f8629ece`KEGGPathway", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/81066f03-423e-4566-800b-da9d03968a36"]|>, {ResourceSystemBase -> "https://www.wolframcloud.com/obj/resourcesystem/api/1.0"}]]["hsa", "00030", "Graph",
VertexSize -> Medium,
VertexStyle -> vertx,
EdgeStyle -> edges]](https://www.wolframcloud.com/obj/resourcesystem/images/beb/bebabad2-b1d9-4b1c-805e-bc3c54c4417a/04f6139a7f9431aa.png) |
| Out[19]= | 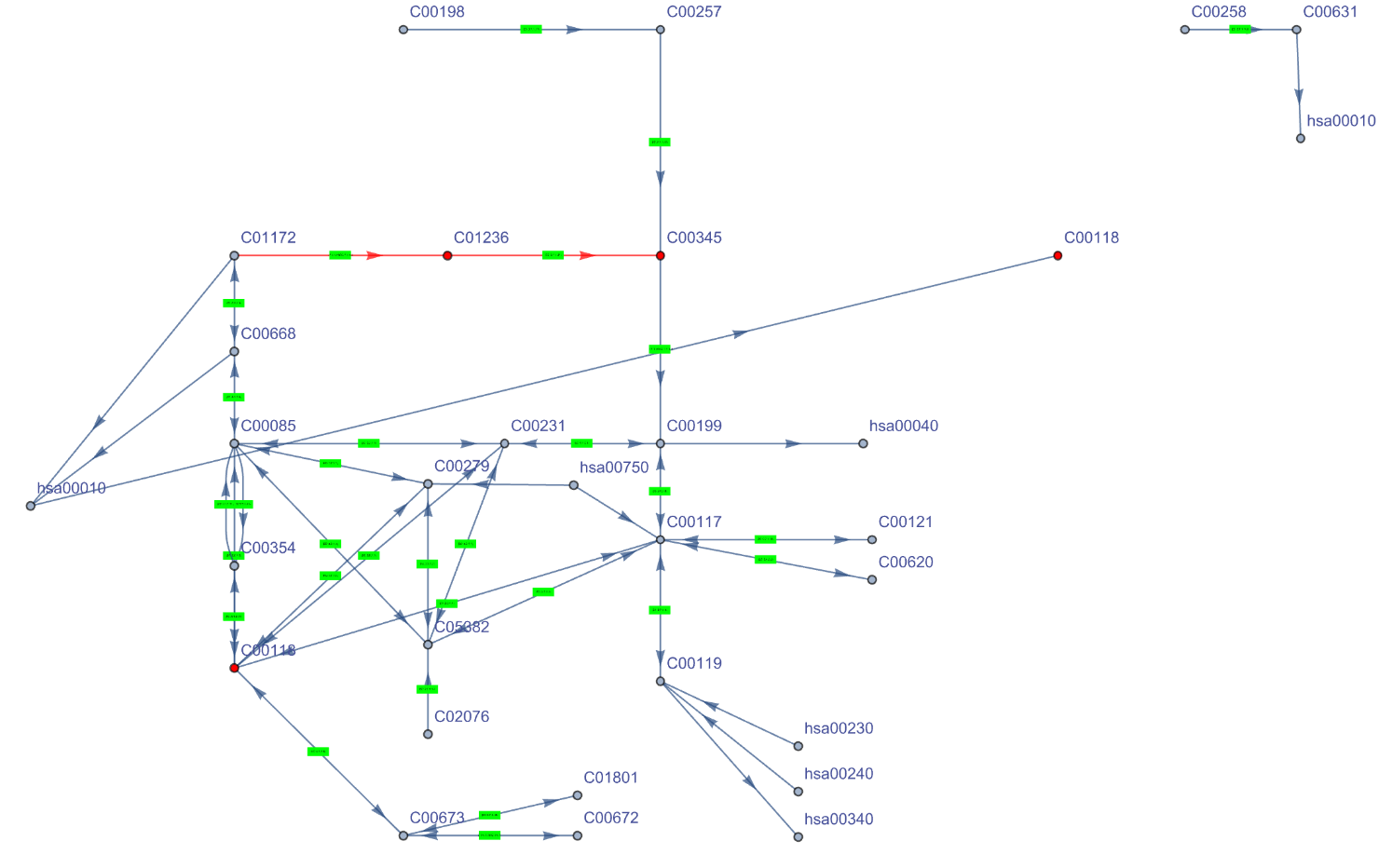 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License