Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get information about a KEGG genome
ResourceFunction["KEGGGenome"]["Genomes"] gives a Dataset with basic information for all the KEGG Genomes. | |
ResourceFunction["KEGGGenome"][keggcode] gives a Dataset with all the properties for a specific keggcode. | |
ResourceFunction["KEGGGenome"][keggcode,prop] gives a property prop for a specific keggcode. | |
ResourceFunction["KEGGGenome"][keyword,"Query"] gives a Dataset of genomes associated with a species-related keyword. |
| "Entry" | ExternalIdentifier of the Genome KEGG code |
| "OrgCode" | organism KEGG code |
| "Category" | designation of KEGG Reference genome and Type strain |
| “Fullname" | the Geome full name |
| “TaxonomicSpecies" | "TaxonomicSpecies" Entity |
| “Annotation" | whether the genome is annotated or not |
| “AnnotationLink" | more information about annotation |
| “Taxonomy" | taxonomy information taken from the NCBI taxonomy database |
| “Lineage" | genetic ancestry or evolutionary path |
| “LinkTaxonomy" | more information about taxonomy |
| “LinkGenomeBrowser" | more information about genome |
| “DataSource" | links to the data source, usually RefSeq. |
| “OriginalDB" | links to the original database where the sequencing was done |
| "Keywords" | associated keywords |
| “Brite" | associated KEGG brite |
| “BriteLink" | associated KEGG brite link |
| “Disease" | disease information for pathogen genomes |
| “Comment" | comment associated |
| “Chromosome" | chromosome information |
| “Plasmid" | plasmid information |
| “Created" | year of creation |
| “Statistics" | statistics of the complete genome |
| “Reference" | references reporting the complete genome (or chromosome) with links to PubMed |
Get the list of all the genomes in the KEGG genome database:
| In[1]:= |
| Out[1]= | 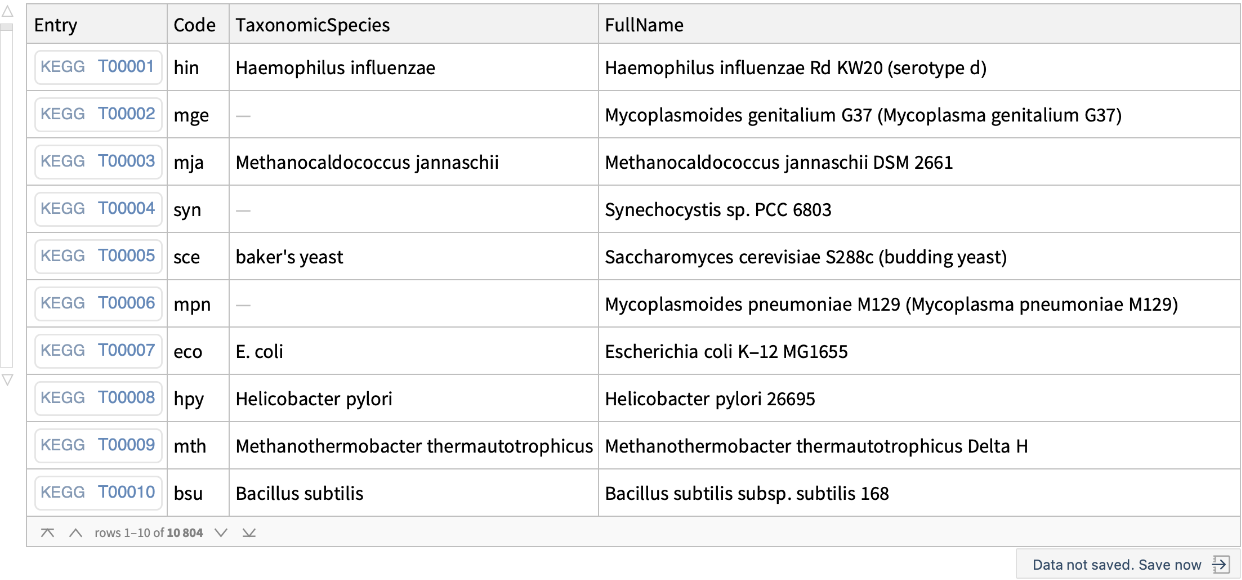 |
Get the information from KEGG about a specific genome:
| In[2]:= |
| Out[2]= | 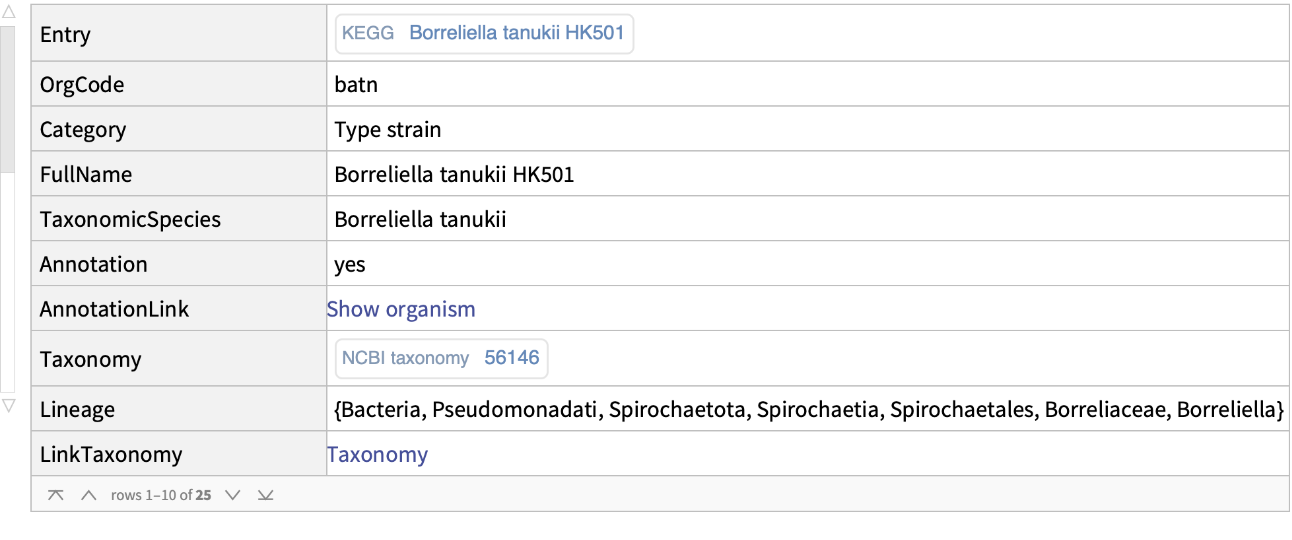 |
Get the KEGG genome codes for a specific query:
| In[3]:= |
| Out[3]= |  |
Keywords may have more than one word:
| In[4]:= |
| Out[4]= |  |
Get the dataset properties independently for each module:
| In[5]:= |
| Out[5]= | 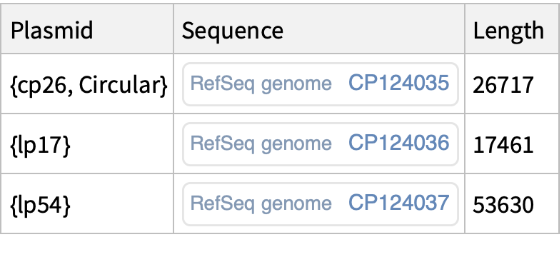 |
You can't give just anything:
| In[6]:= |
| Out[6]= |
Only valid KEGG codes are supported:
| In[7]:= |
| Out[7]= |
Unrecognized queries give an error:
| In[8]:= |
| Out[8]= |
Retrieve the "Number of nucleotides", "Number of protein genes", and "Number of RNA genes" for various species, grouped by phylum:
| In[9]:= |
| In[10]:= | ![SeedRandom[1];
sampleSpe = RandomChoice[txSpecies, 100];
phylum = # -> #[EntityProperty["TaxonomicSpecies", "Phylum"]] & /@ sampleSpe;
moreFrePhy = Select[Tally[phylum[[All, 2]]], #[[2]] > 4 &];](https://www.wolframcloud.com/obj/resourcesystem/images/625/625605ce-e351-4851-8cdf-b86968a490ee/26566cdb7a4b36b9.png) |
| In[11]:= | ![sampleSpe2 = entriesANDSp[
Select[
MemberQ[sampleSpe, #TaxonomicSpecies] &&
MemberQ[moreFrePhy[[All, 1]], ReplaceAll[#TaxonomicSpecies, phylum]]
&]
][All, Join[#, <|"Phylum" -> ReplaceAll[#TaxonomicSpecies, phylum]|>] &];](https://www.wolframcloud.com/obj/resourcesystem/images/625/625605ce-e351-4851-8cdf-b86968a490ee/4bde0896899c0cf4.png) |
| In[12]:= |
| In[13]:= |
Visualize the results:
| In[14]:= | ![BoxWhiskerChart[
Map[ToExpression, DeleteMissing[
Lookup[Normal[#[[All, 2]]], "NumberOfNucleotides"]]] & /@ sampleSpe3Grp,
ChartLabels -> Automatic, BarOrigin -> Left, FrameLabel -> {"Number of nucleotides"}]](https://www.wolframcloud.com/obj/resourcesystem/images/625/625605ce-e351-4851-8cdf-b86968a490ee/7f1b9eec18db643a.png) |
| Out[14]= | 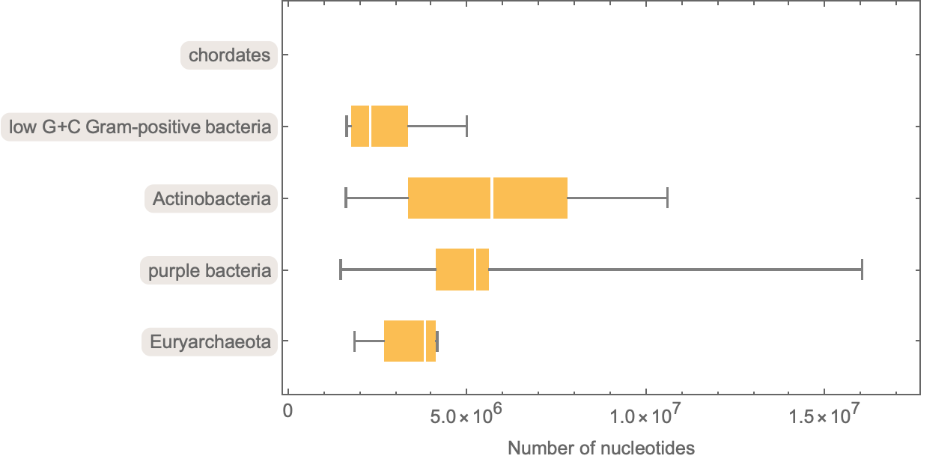 |
| In[15]:= | ![BoxWhiskerChart[
Map[ToExpression, DeleteMissing[
Lookup[Normal[#[[All, 2]]], "NumberOfProteinGenes"]]] & /@ sampleSpe3Grp,
ChartLabels -> Automatic, BarOrigin -> Left, FrameLabel -> {"Number of protein genes"}]](https://www.wolframcloud.com/obj/resourcesystem/images/625/625605ce-e351-4851-8cdf-b86968a490ee/2579d31e77ac6851.png) |
| Out[15]= |  |
| In[16]:= | ![BoxWhiskerChart[
Map[ToExpression, DeleteMissing[
Lookup[Normal[#[[All, 2]]], "NumberOfRNAGenes"]]] & /@ sampleSpe3Grp,
ChartLabels -> Automatic, BarOrigin -> Left, FrameLabel -> {"Number of RNA genes"}]](https://www.wolframcloud.com/obj/resourcesystem/images/625/625605ce-e351-4851-8cdf-b86968a490ee/177ed13f1d5a044c.png) |
| Out[16]= | 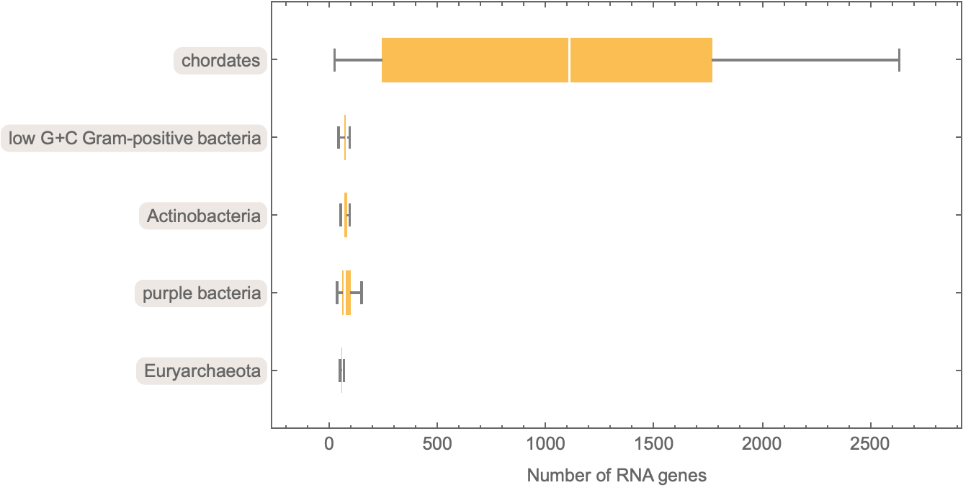 |
Wolfram Language 13.0 (December 2021) or above
This work is licensed under a Creative Commons Attribution 4.0 International License