Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get biotic interactions of a specified taxon using the Global Biotic Interactions (GloBI) API
ResourceFunction["GloBIData"][taxon] gives a Graph of taxa that the specified taxon interacts with. | |
ResourceFunction["GloBIData"][taxon,"Dataset"] gives a Dataset containing information about the interactions of the specified taxon. |
| "InteractionType" | "InteractsWith" | interaction types to return |
| "TargetTaxon" | "NotSpecified" | target taxon interacting with source taxon |
| "IncludeObservations" | True | whether to include interaction observations |
| "Eats" | "EatenBy" | "PreysOn" |
| "PreyedUponBy" | "Kills" | "KilledBy" |
| "ParasiteOf" | "HasParasite" | "EndoparasiteOf" |
| "HasEndoparasite" | "EctoparasiteOf" | "HasEctoparasite" |
| "ParasitoidOf" | "HasParasitoid" | "HostOf" |
| "HasHost" | "Pollinates" | "PollinatedBy" |
| "PathogenOf" | "HasPathogen" | "VectorOf" |
| "HasVector" | "DispersalVectorOf" | "HasDispersalVector" |
| "SymbiontOf" | "MutualistOf" | "CommensalistOf" |
| "FlowersVisitedBy" | "VisitsFlowersOf" | "EcologicallyRelatedTo" |
| "CoOccursWith" | "InteractsWith" |
Create a taxonomic Graph from the distinct prey taxa of a specific taxon:
| In[1]:= |
| Out[1]= | 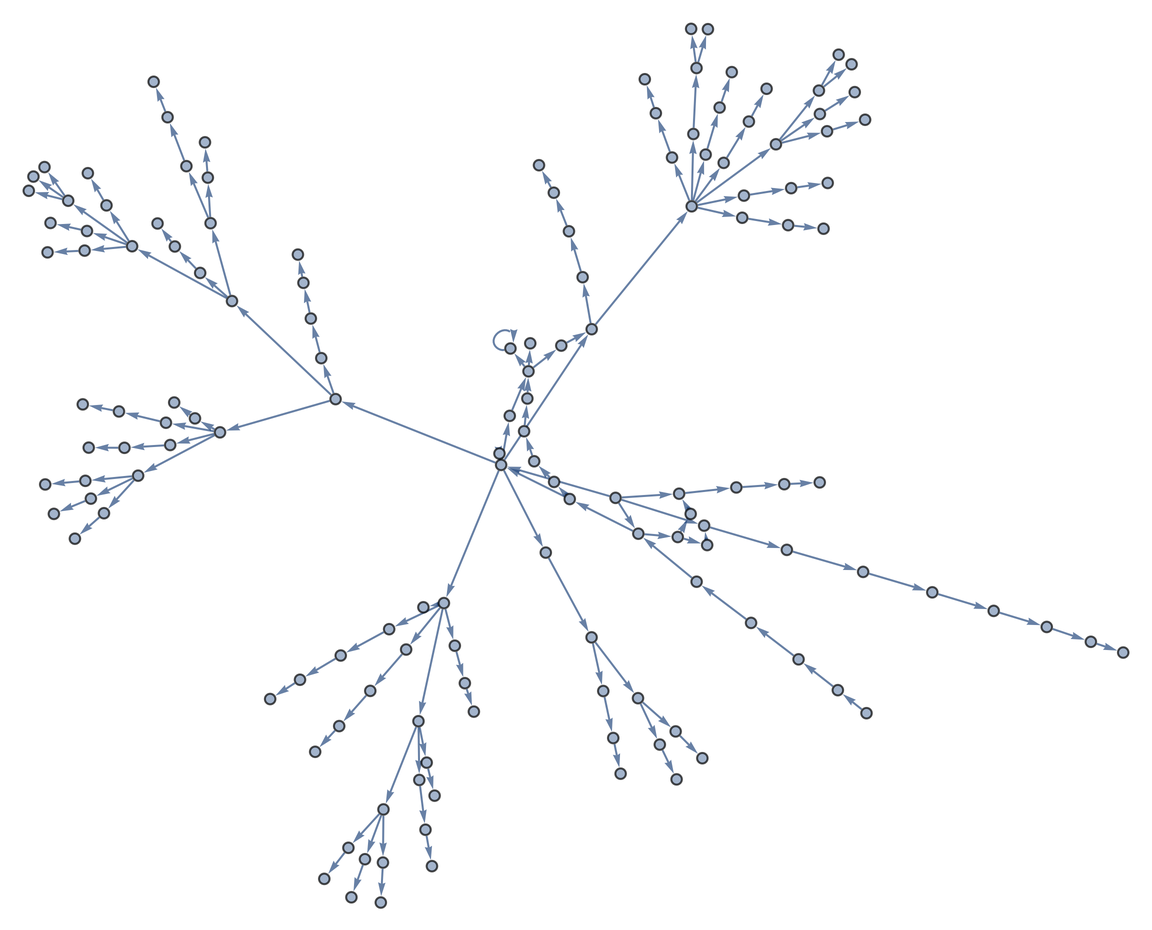 |
Create a Graph from all interaction observations between specific taxa and specific interaction type:
| In[2]:= | ![ResourceFunction["GloBIData"][Entity["Species", "Species:CanisLupus"],
"InteractionType" -> "Eats", "TargetTaxon" -> Entity["Species", "Class:Aves"]]](https://www.wolframcloud.com/obj/resourcesystem/images/740/74041c3a-f659-4059-bba3-258cfc0b24f4/769c70a232006163.png) |
| Out[2]= | 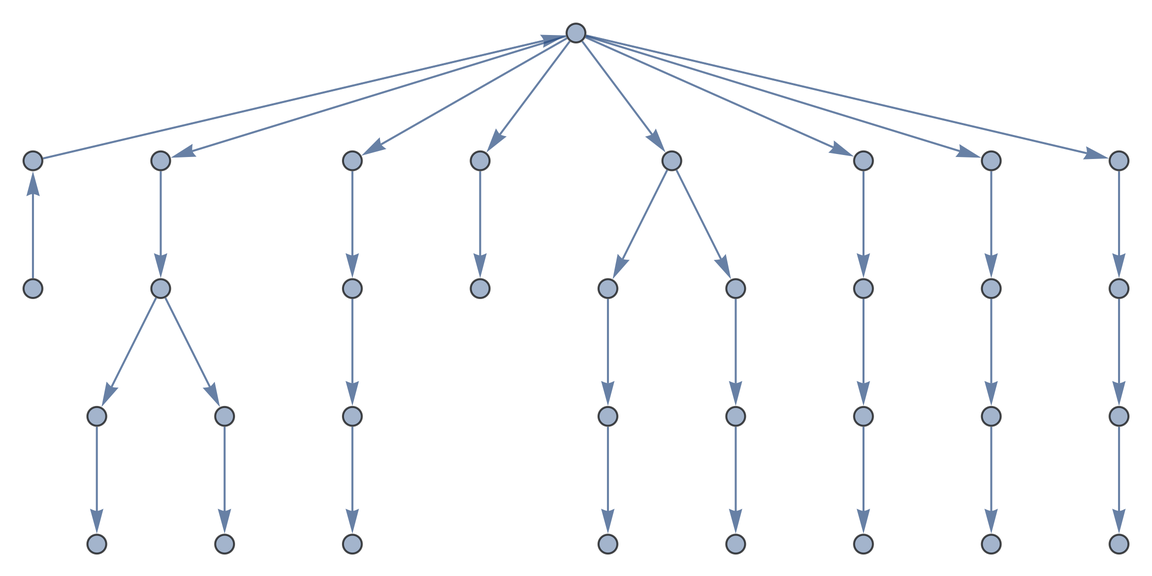 |
Obtain a Dataset with all interaction observations specified by taxa and interaction type:
| In[3]:= | ![ResourceFunction["GloBIData"][
Entity["Species", "Species:CanisLupus"], "Dataset", "InteractionType" -> "Eats", "TargetTaxon" -> Entity["Species", "Family:Cervidae"]]](https://www.wolframcloud.com/obj/resourcesystem/images/740/74041c3a-f659-4059-bba3-258cfc0b24f4/468240de38f21ad3.png) |
| Out[3]= | 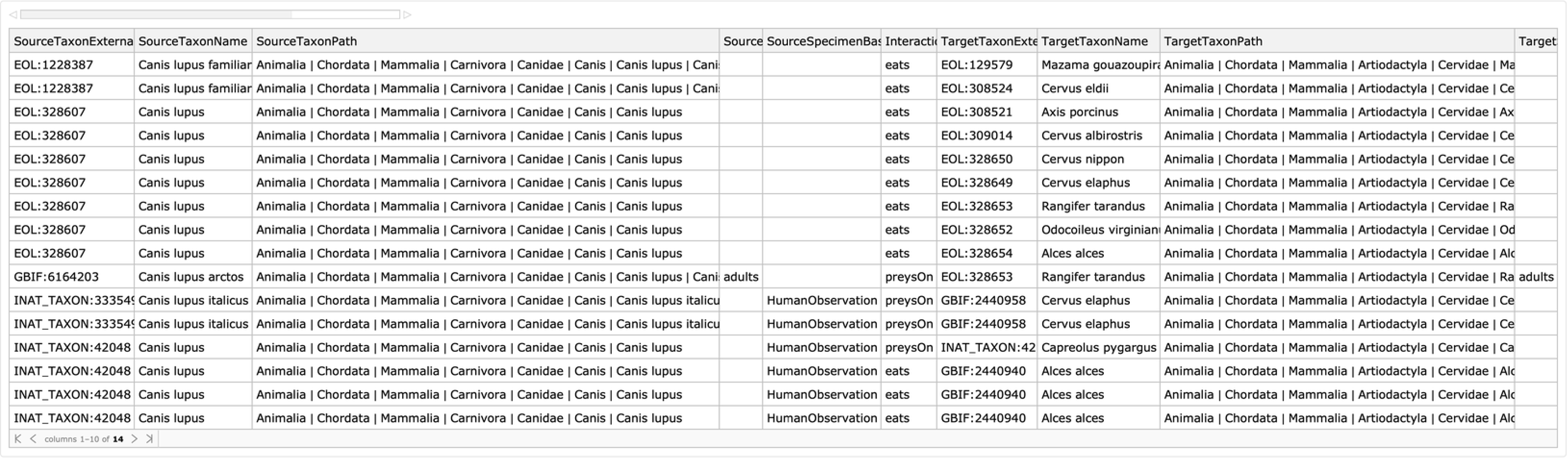 |
Find all parasitic interactions between mites and ticks and their ape hosts:
| In[4]:= | ![ResourceFunction["GloBIData"][
Entity["Species", "Class:Arachnida"], "Dataset", "InteractionType" -> "ParasiteOf", "TargetTaxon" -> Entity["Species", "Family:Hominidae"]]](https://www.wolframcloud.com/obj/resourcesystem/images/740/74041c3a-f659-4059-bba3-258cfc0b24f4/2322be0548463f73.png) |
| Out[4]= | 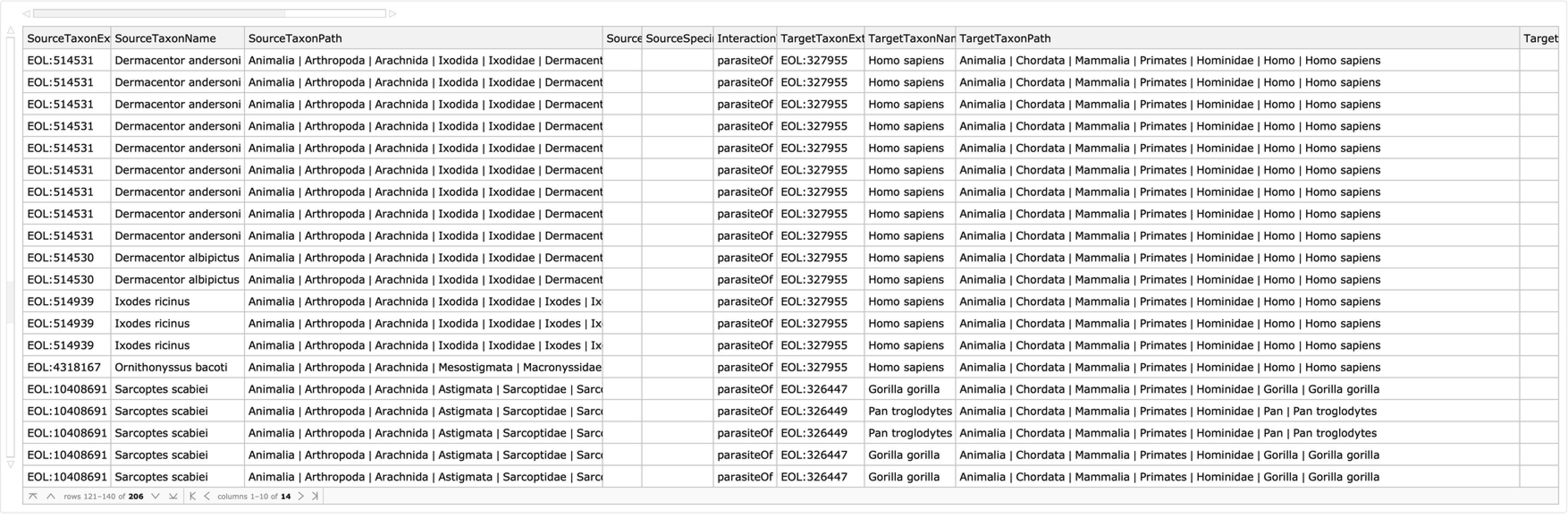 |
Create a food web of several species that interact with each other:
| In[5]:= | ![getSpeciesEatenBy[species_] := Union@Flatten@
StringCases[
Normal@ResourceFunction["GloBIData"][species, "Dataset", "InteractionType" -> "Eats"][[All, "TargetTaxonName"]], __ ~~ " " ~~ __]](https://www.wolframcloud.com/obj/resourcesystem/images/740/74041c3a-f659-4059-bba3-258cfc0b24f4/0d8acbc449bd2fb2.png) |
| In[6]:= | ![speciesEatenByRiverOtter = getSpeciesEatenBy[Entity["Species", "Species:LutraLutra"]];
speciesEatenByTrout = getSpeciesEatenBy["Salmo trutta"];
speciesEatenBySprat = getSpeciesEatenBy["Sprattus sprattus"];](https://www.wolframcloud.com/obj/resourcesystem/images/740/74041c3a-f659-4059-bba3-258cfc0b24f4/5ceef088aef87d32.png) |
| In[7]:= | ![Labeled[Legended[
Graph[Join[
Map[Style["Salmo trutta" -> #, Orange] &, speciesEatenByTrout], Map[Style["Lutra lutra" -> #, Blue] &, speciesEatenByRiverOtter], Map[Style["Sprattus sprattus" -> #, Darker[Green]] &, speciesEatenBySprat]], ImageSize -> Large, VertexLabels -> Placed["Name", Tooltip]],
SwatchLegend[{Blue, Orange, Darker[Green]}, {"River Otter", "Brook Trout", "Sprat"}]], Style["Foodweb of three different species", FontFamily -> "Helvetica", Bold, FontSize -> 18], Top]](https://www.wolframcloud.com/obj/resourcesystem/images/740/74041c3a-f659-4059-bba3-258cfc0b24f4/23b9d19ff976e1cc.png) |
| Out[7]= |  |
This work is licensed under a Creative Commons Attribution 4.0 International License