Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Fold a protein using the ESMFold API
ResourceFunction["ESMFoldProteinSequence"][p] shows a visualization of the protein structure represented by the sequence p predicted using the ESMFold API. |
Visualize the structure of a protein sequence as predicted by ESMFold:
| In[1]:= |
| Out[1]= | 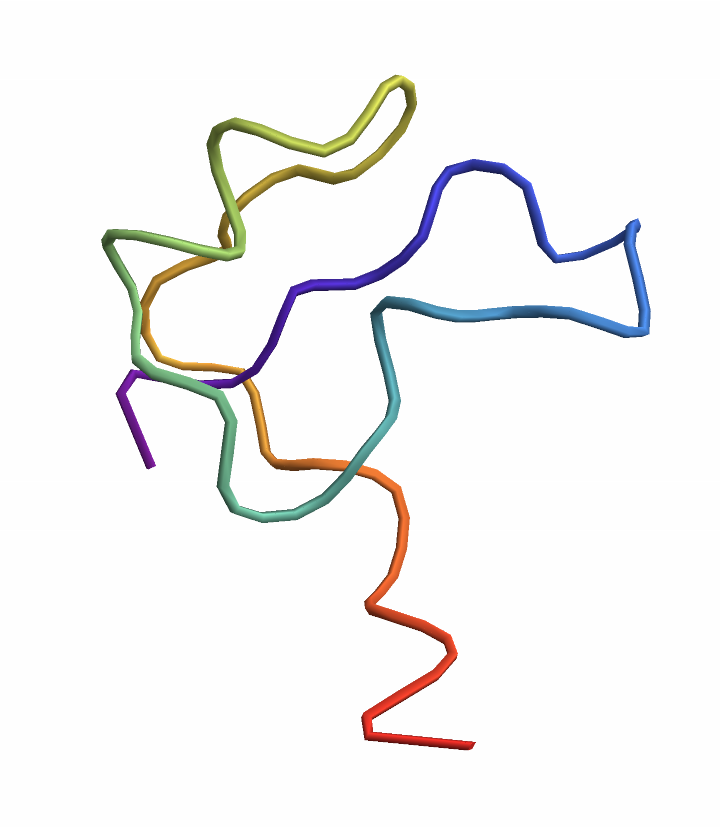 |
Visualize the folding of a peptide represented as a BioSequence:
| In[2]:= | ![ResourceFunction["ESMFoldProteinSequence"][
BioSequence[
"Peptide", "GSGHMDKKYSIGLAIGTNSVGWAVITDEYKVPSKKFKVLGNTDRHSIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRILYLQEIFSNEMAKVDDSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLVDSTDKADLRLIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINASGVDAKAILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKDTYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDEHHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKMDGTEELLVKLNRE", {}]]](https://www.wolframcloud.com/obj/resourcesystem/images/1b5/1b5b740a-25b3-4106-99e4-be742f162e69/55d6c010d5fb13f7.png) |
| Out[2]= | 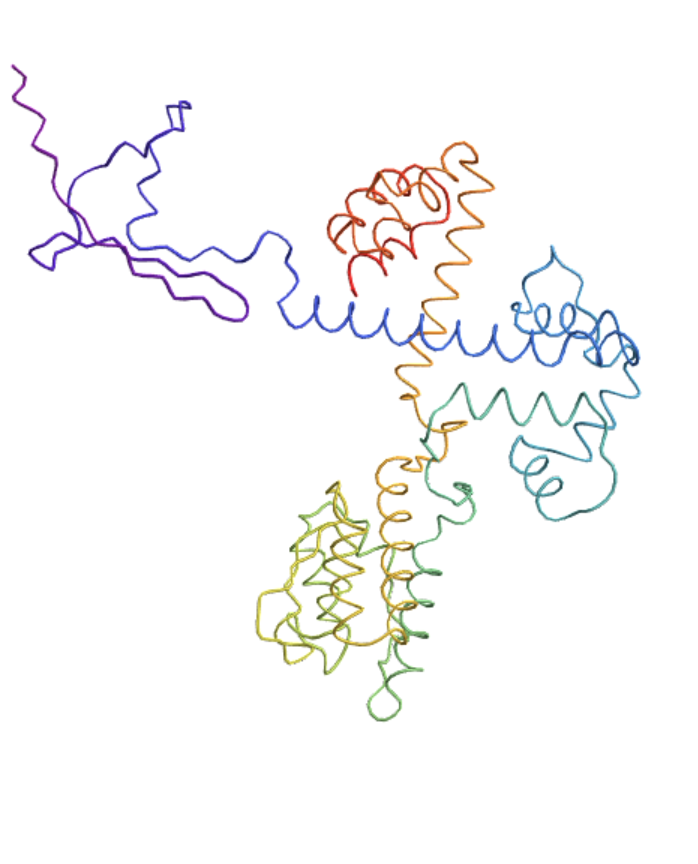 |
Predict the structure of a protein Entity:
| In[3]:= |
| Out[3]= | 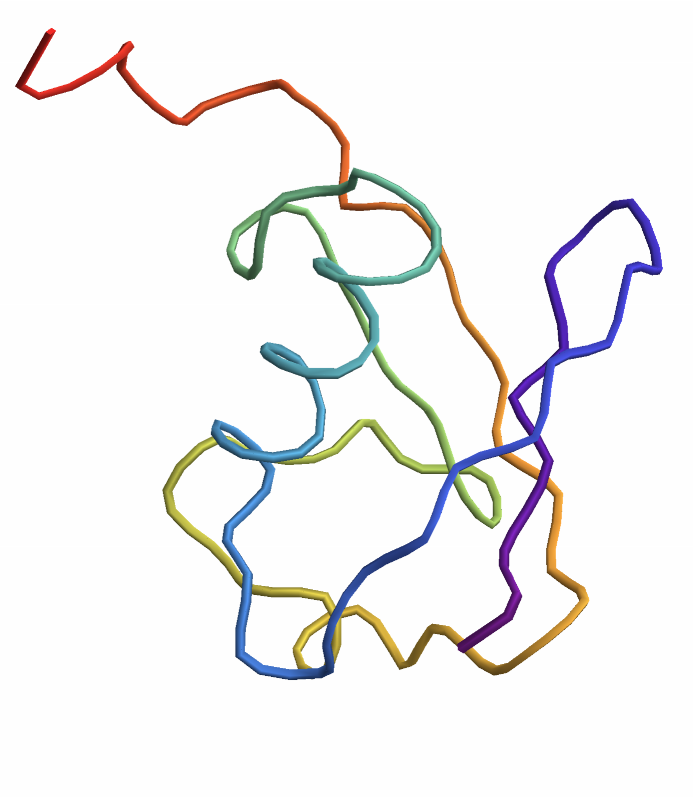 |
Compare with the known structure:
| In[4]:= | ![EntityValue[Entity["Protein", "NEDD8"], EntityProperty["Protein", "StructureDiagram"]]](https://www.wolframcloud.com/obj/resourcesystem/images/1b5/1b5b740a-25b3-4106-99e4-be742f162e69/49ee7dcaf26d167d.png) |
| Out[4]= | 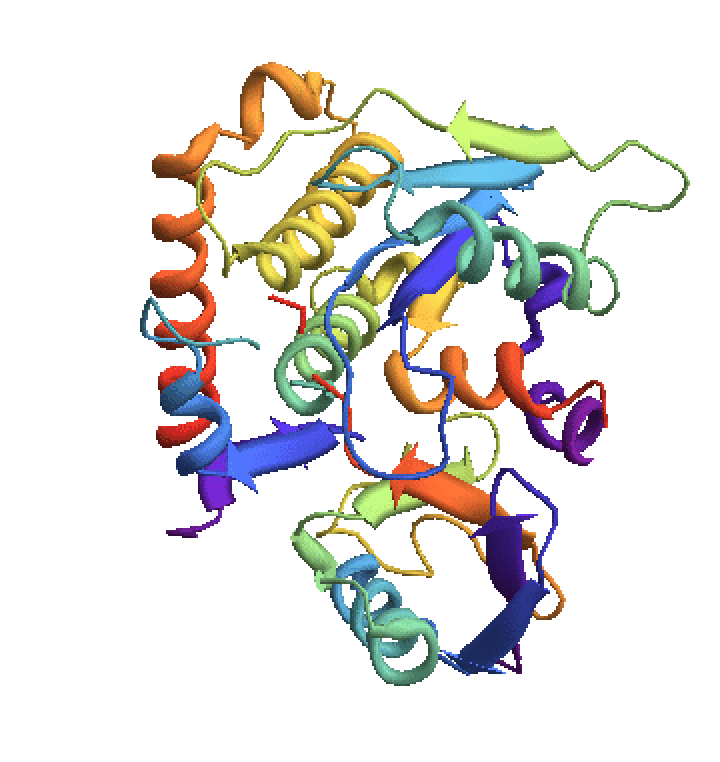 |
This work is licensed under a Creative Commons Attribution 4.0 International License