Basic Examples (12)
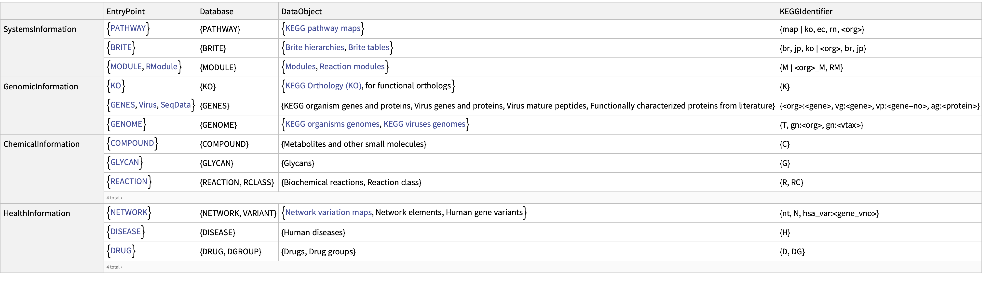
Get the list of all the KEGG databases and additional information:
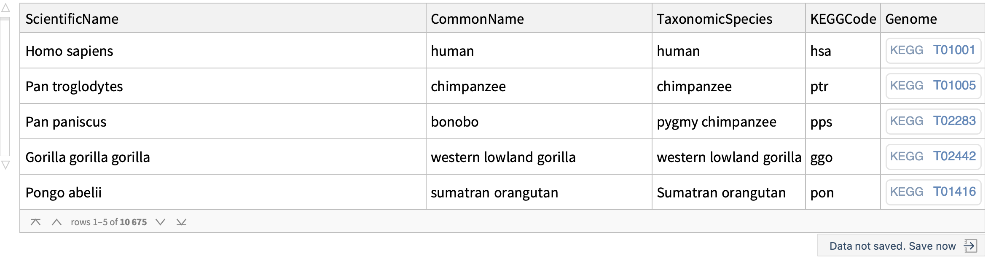
Get the list of all the organisms in KEGG with their KEGG code and genome entry:
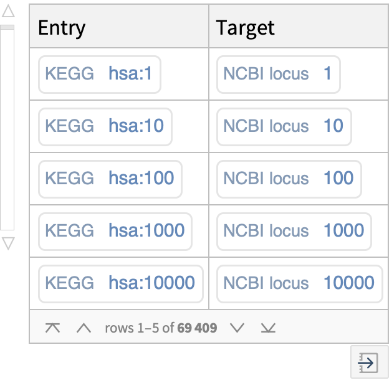
Retrieve, for each human gene in KEGG, the associated entry identifiers (accession numbers) from external databases (i.e., NCBIGeneID, NCBIProteinID, and UniProt):
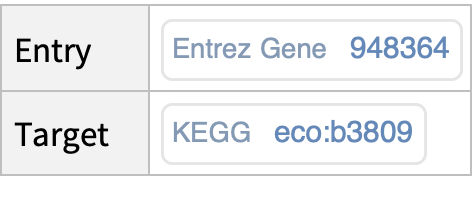
Retrieve, for a specific human gene in KEGG, the associated entry identifiers (accession numbers) from external databases (i.e., NCBIGeneID, NCBIProteinID, UniProt):
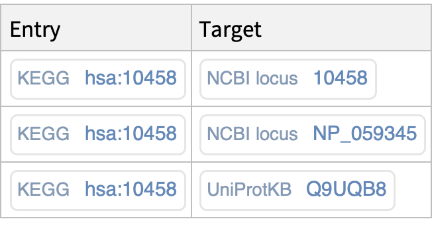
Retrieve, for a specific identifier of outside database (i.e., NCBIGeneID, NCBIProteinID, UniProt), the associated KEGG entry identifier:
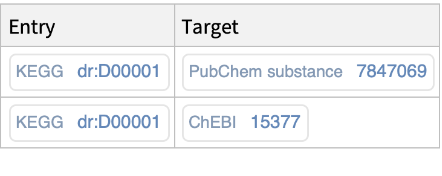
Retrieve, for a specific KEGG chemical substance identifier (i.e., Compound, Glycan, or Drug), the associated entry identifiers (accession numbers) from external databases (i.e., PubChem, ChEBI):
Retrieve, for a specific identifier of outside database (i.e.,PubChem, ChEBI), the associated KEGG chemical substance identifiers (i.e., Compound, Glycan, Drug):
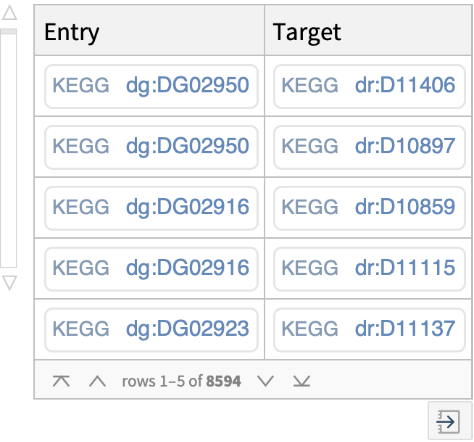
Get cross-references between KEGG DGroup database and all KEGG databases and some outside databases (i.e., PubMed, ATC, JTC, NDC, YK):
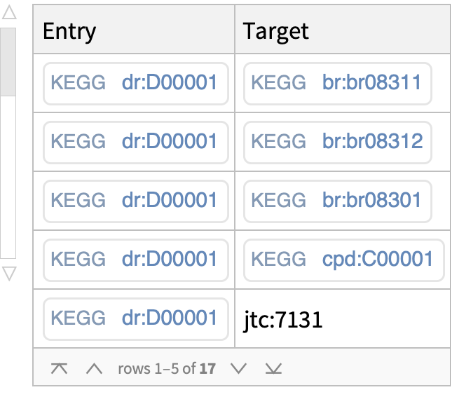
Get cross-references between a specific KEGG entry and all KEGG databases including some outside databases (i.e., PubMed, ATC, JTC, NDC, YK):
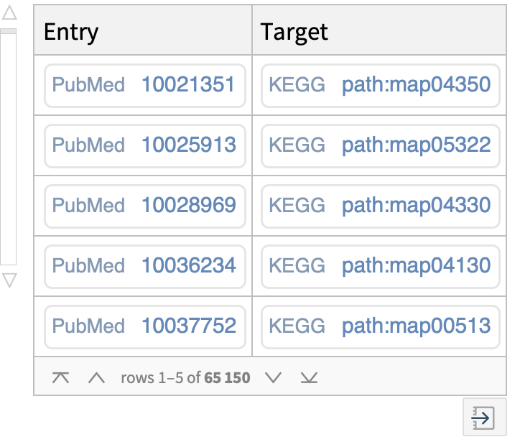
Find various relationships or cross-references between an external database (i.e., PubMed, ATC, JTC, NDC, YK) and KEGG databases:
Get cross-references between an outside database entry (i.e., PubMed, ATC, JTC, NDC, YK) and all KEGG databases:
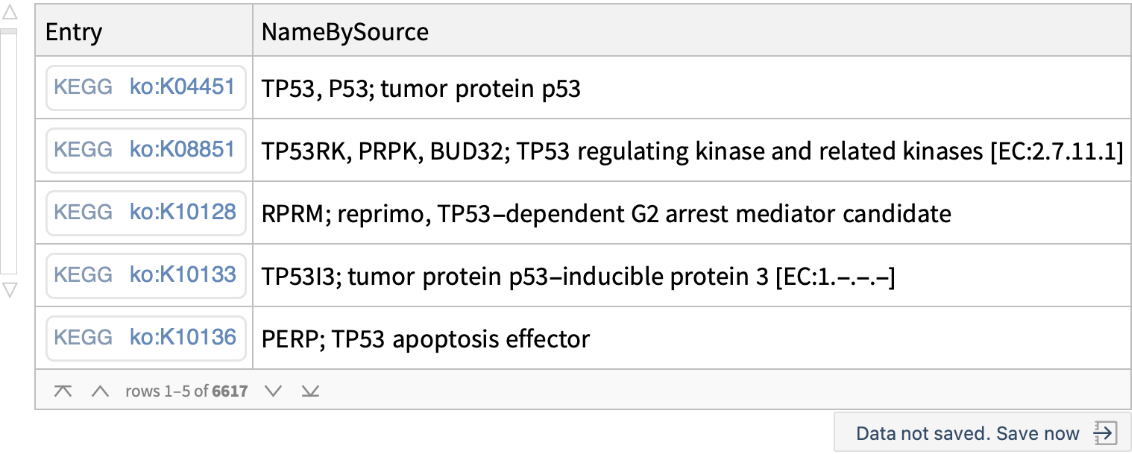
Find all the entries matching query keyword "tp53":
Scope (2)
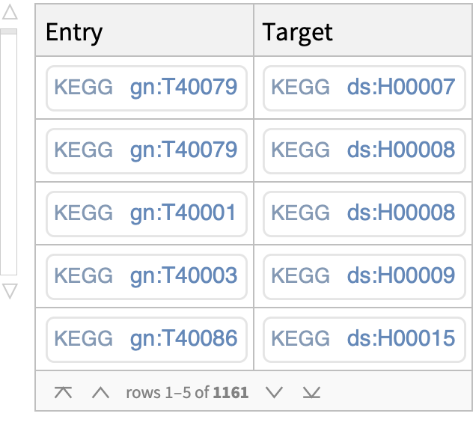
Retrieve, for a list of human genes in KEGG, the associated entry identifiers (accession numbers) from external databases (i.e., NCBIGeneID, NCBIProteinID, UniProt):
For a list of selected KEGG gene identifiers, get cross-references within all KEGG databases and some outside databases (i.e., PubMed, ATC, JTC, NDC, YK):
Options (11)
Retrieve, for each gene in KEGG human genome (i.e., T01001), the associated entry identifiers (accession numbers) from a specific external database:
Retrieve, for all the entries in a chemical substance KEGG database (i.e., Compound, Glycan, Drug), the associated entry identifiers (accession numbers) from a specific external database:
Retrieve, for a specific human gene in KEGG, the associated entry identifiers (accession numbers) from a specific external database:
Retrieve, for a specific identifier of outside database (i.e., NCBIGeneID, NCBIProteinID, UniProt), the associated KEGG gene entry:
Retrieve, for a specific KEGG chemical substance identifier (i.e., Compound, Glycan, Drug), the associated entry identifiers of PubChem outside database:
Get KEGG gene database to KEGG disease database cross-references or the relationships between them:
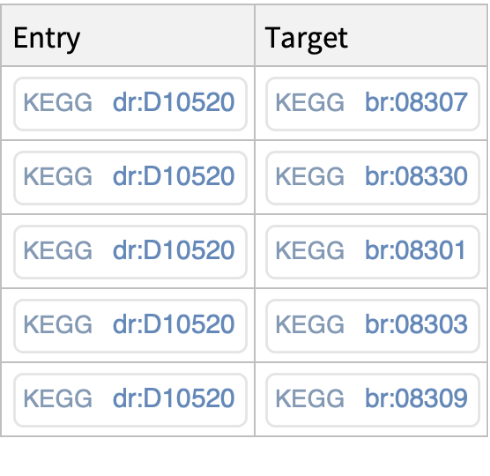
For a selected entry, get cross-references with the KEGG brite database:
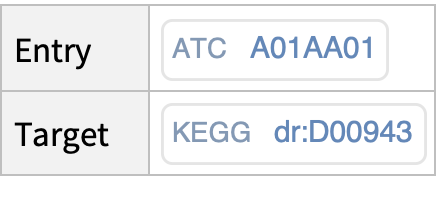
Get cross-references between an outside database entry (i.e., PubMed, ATC, JTC, NDC, YK) and the drug KEGG database:
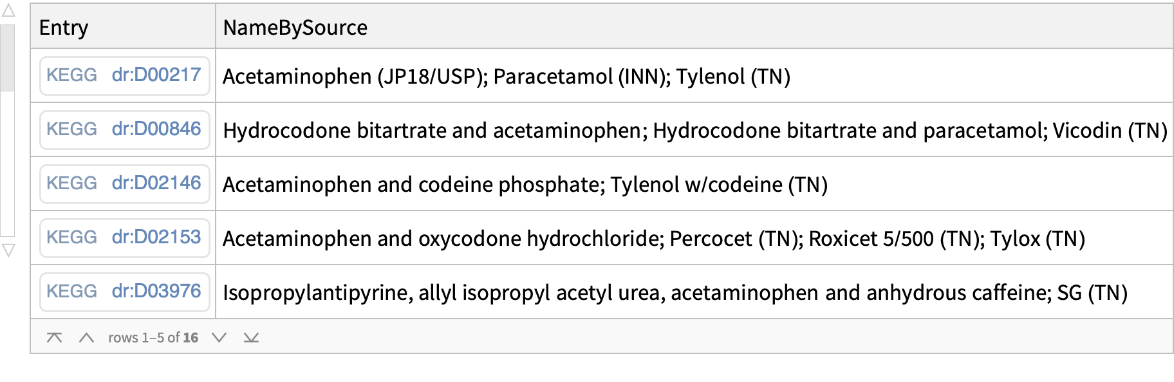
Find all the KEGG Drug entries matching the query keyword acetaminophen:
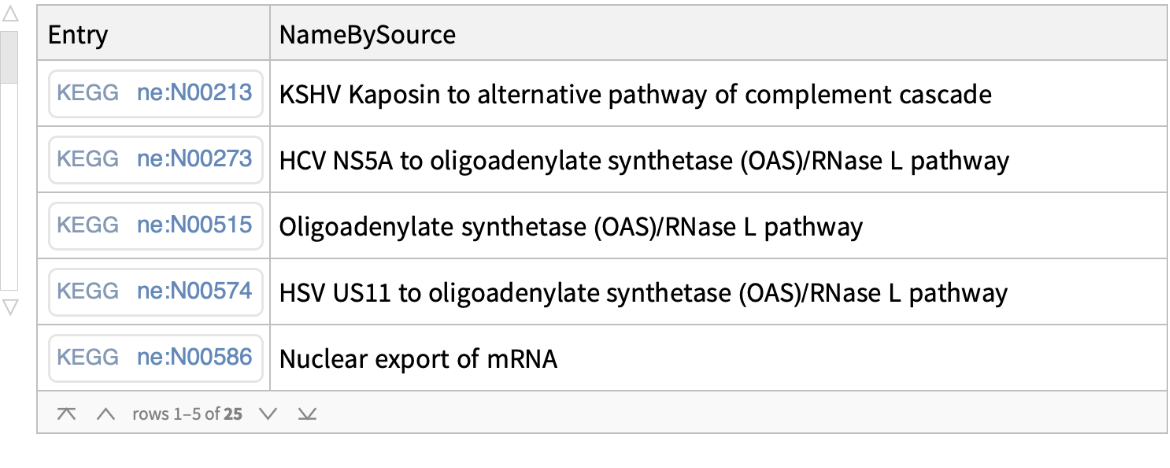
Find all the KEGG Network and Pathway entries matching query keyword RNA:
Applications (3)
Get the compounds in the KEGG Pathway map00020 using the current function:
Get the compounds in the KEGG Pathway map00020 using KEGGPathway:
Compare both lists to check if they contain the same compounds:
Possible Issues (1)
You get Missing when the entry is not correct:
Neat Examples (2)
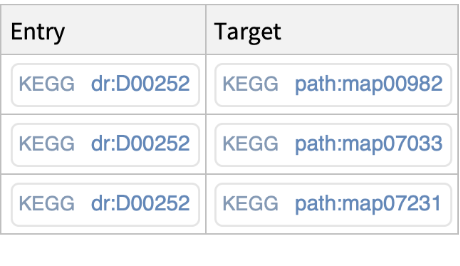
Get all the KEGG pathways associated with the D00252 drug:
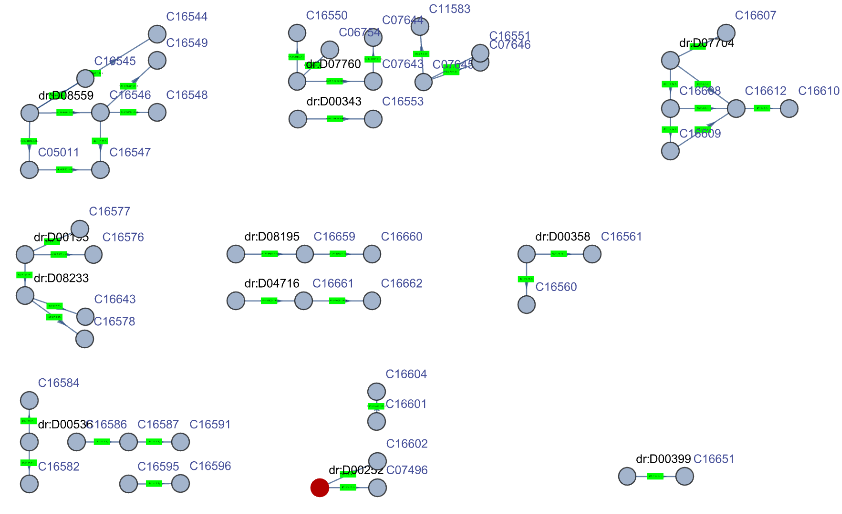
Get a Graph of one of the pathways and highlight the D00252 drug:






![crossRefe = ResourceFunction["KEGGReference"]["map00020", "KEGGCrossReference", "Database" -> {"compound", "ko"}];
cpdCrossRef = Normal[crossRefe[Select[StringContainsQ[#Target[[2]], "cpd"] &]][All,
2]][[All, 2]]](https://www.wolframcloud.com/obj/resourcesystem/images/618/61899202-156e-4c7d-b7b8-36a938bd3a08/34495707cb1fe73c.png)