Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Search the PubChem database for similar compounds
ResourceFunction["PubChemSimilaritySearch"][mol] returns a list of "PubChemCompoundID" identifiers for compounds similar to the molecule or chemical entity mol. | |
ResourceFunction["PubChemSimilaritySearch"][mol,"Molecule"] returns a list of Molecule objects constructed from the external identifiers similar to mol. |
| "Similarity2DSearch" | Tanimoto similarity of topological fingerprints |
| "Similarity3DSearch" | Tanimoto similarity of 3D shape fingerprints |
| "Original" | exact match to input |
| "Parent" | parent compound |
| "SameStereo" | same stereo |
| "SameIsotopes" | same isotopes |
| "SameConnectivity" | same connectivity |
| "SameFormula" | same molecular formula |
| "SameTautomer" | same tautomer |
| "SameParent" | same parent |
| "SameParentStereo" | same parent stereo |
| "SameParentIsotopes" | same parent isotopes |
| "SameParentConnectivity" | same parent connectivity |
| "SameParentTautomer" | same parent tautomer |
Find the compound ID for a similar molecules:
| In[1]:= |
| Out[1]= | 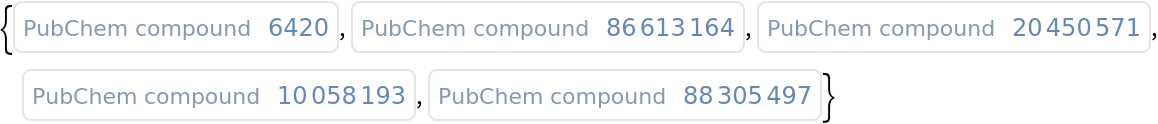 |
Perform the same search, but return the results as Molecule objects:
| In[2]:= |
| Out[2]= |  |
Visualize the results using MoleculePlot:
| In[3]:= |
| Out[3]= | 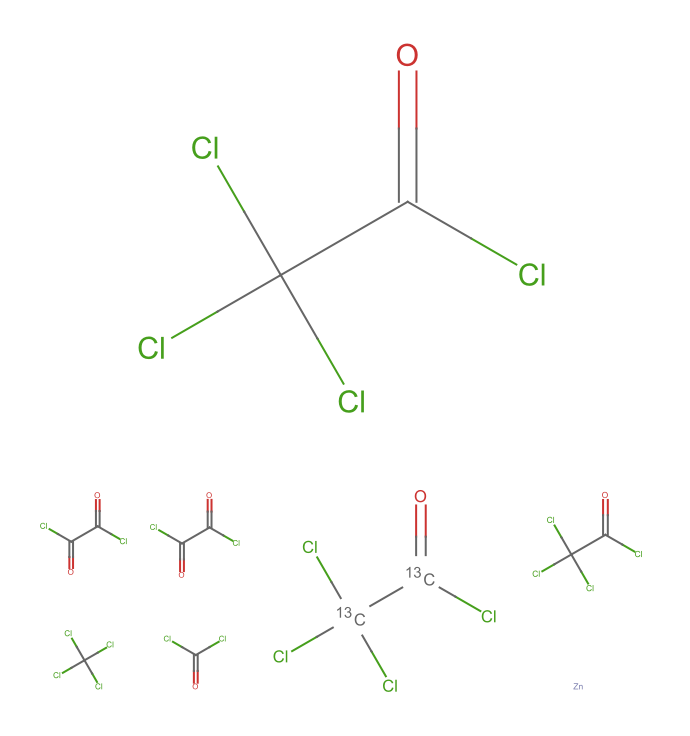 |
Search PubChem for tautomers:
| In[4]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/9a4428cd-ea41-47a7-860e-a16a9366c3e5"]](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/611941fc20adfe3f.png) |
| Out[4]= |
Get a list of molecules with the same connectivity as adenosine:
| In[5]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/95babadb-167b-45b8-bfe7-cf89d2ef832b"]](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/26b0545890defe3c.png) |
| Out[5]= |  |
Visualize the molecules using MoleculePlot:
| In[6]:= |
| Out[6]= | 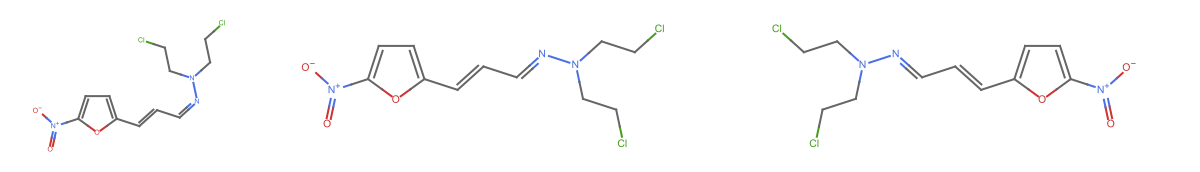 |
Get a list of IDs for compounds with the same parent:
| In[7]:= | ![ResourceFunction["PubChemSimilaritySearch"][
Molecule["O[C@H](c1cc(nc2c1cccc2C(F)(F)F)C(F)(F)F)[C@H]1CCCCN1"], "SearchType" -> "SameParent"]](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/242f21dd8b49e468.png) |
| Out[7]= |
Get a list of isomers by using the "SameFormula" search type:
| In[8]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/0a669263-c091-49a9-8062-6aa34081dbbf"]](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/0bfc446c9d0aefc6.png) |
| Out[8]= | 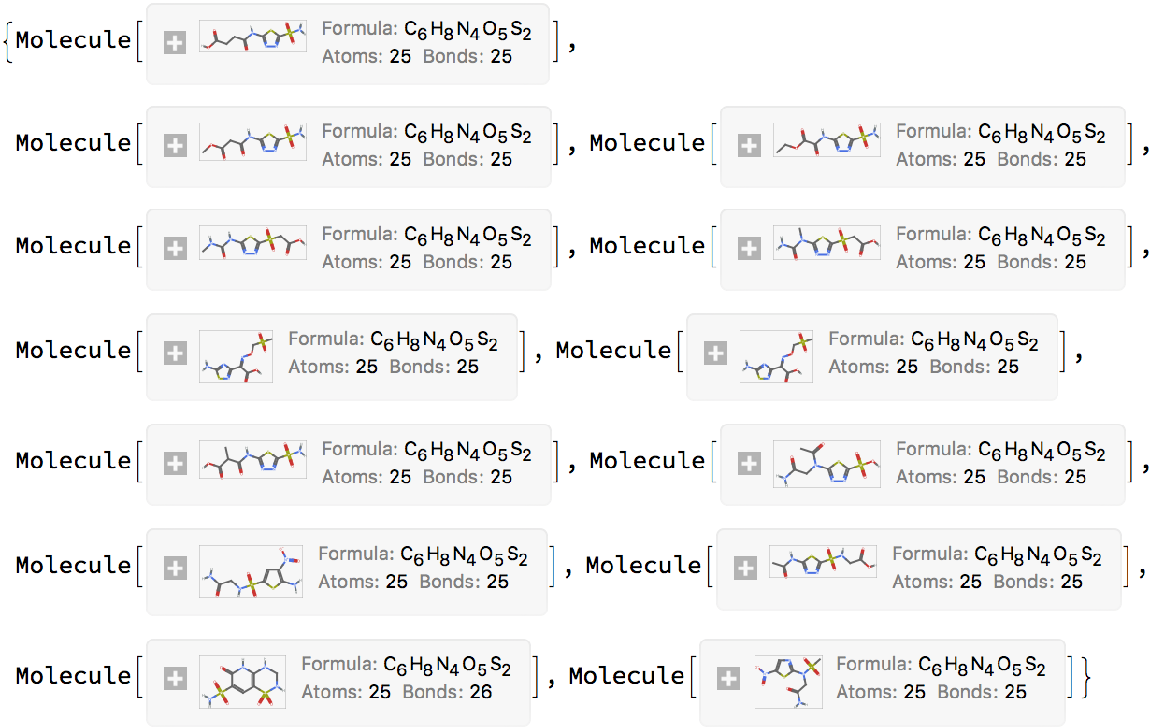 |
PubChemSimilaritySearch can be used with Entity or Molecule objects:
| In[9]:= | ![ResourceFunction["PubChemSimilaritySearch"][#, "SearchType" -> "Original"] & /@ {Molecule["caffeine"], Entity["Chemical", "Caffeine"]}](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/5b97dc7552aa18fa.png) |
| Out[9]= |
PubChemSimilaritySearch will automatically thread over lists:
| In[10]:= | (C)C)O", "O=C1CC[C@]2(C(=C1)CC[C@@H]1[C@@H]2[C@@H](O)C[C@]2([C@H]1CC[C@]2(O)C(=O)COS(=O)(=O)C)C)C", "CCCP(C1(C)CCC1)C", "SC1CC1", "COC(=O)c1cccnc1", "O=C1CC[C@@H]2[C@]1(C)CC[C@H]1[C@H]2CCc2c1ccc(c2)OS(=O)(=O)N", "CC[C@@H](C(=O)O[C@@H]1CC(C)(C)C[C@H]2[C@]1(CC[C@@]1(C2=CC[C@H]2[C@@]1(C)CC[C@@H]1[C@]2(C)CCC(=O)C1(C)C)C)C(=O)O)C", "CC[C@H](N[C@H]1CC[C@@H]1C)C", "CC[C@H]1OC(=O)[C@H](C)[C@@H](O[C@@H]2O[C@@H](C)[C@@H]([C@](C2)(C)OC)O)[C@H](C)[C@@H](O[C@@H]2O[C@H](C)C[C@@H]([C@H]2O)N(C)C)[C@](C[C@H](CN([C@@H]([C@H]([C@]1(C)O)O)C)C)C)(C)O", "CCOc1nc(N)nc2c1ncn2[C@@H]1C[C@@H]([C@H](O1)CO)O"};
Length@ResourceFunction["PubChemSimilaritySearch"][mols]](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/2f45b39f38f1e372.png) |
| Out[11]= |
By adjusting the Tanimoto threshold, the number of similar compounds returned can be controlled:
| In[12]:= | ![mol = Molecule[
"CCOC(=O)C1=C(C)NC(=C([C@@H]1c1c(F)c(F)c(c(c1F)F)F)C(=O)OCC)C"];
similarityData = ResourceFunction[
"DynamicMap"][{#, Length@ResourceFunction["PubChemSimilaritySearch"][mol, "TanimotoThreshold" -> #]} &,
Join[Range[60, 90, 10], Range[91, 99]]]](https://www.wolframcloud.com/obj/resourcesystem/images/59b/59b2fd6c-be0b-4724-8a28-5e2475f7e343/75f911da6e5dbe5a.png) |
| Out[12]= |
Use ListLogPlot to visualize the relationship between threshold and the size of the similarity space:
| In[13]:= |
| Out[13]= | 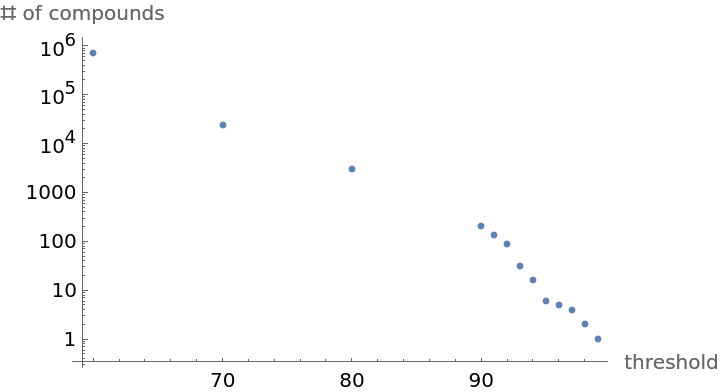 |
This work is licensed under a Creative Commons Attribution 4.0 International License