Wolfram Function Repository
Instant-use add-on functions for the Wolfram Language
Function Repository Resource:
Get a random SMILES string for a molecule
ResourceFunction["RandomSmilesString"][mol] returns a SMILES string for the molecule mol with the atoms in a random order. | |
ResourceFunction["RandomSmilesString"][mol,n] returns a list of n SMILES strings. |
| "AllBondsExplicit" | False | whether to explicitly show all bonds |
| "AllHsExplicit" | False | whether to explicitly list all implicit hydrogens |
| "Isomeric" | True | include stereochemistry and isotope information |
| "Kekulized" | False | whether to use aromatic or Kekule form |
| IncludeHydrogens | False | whether to include hydrogens as explicit atoms |
Create a molecule and get a random SMILES string:
| In[1]:= | ![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/bb62831f-99af-4f97-a8b7-91d5b4b3e4ba"]](https://www.wolframcloud.com/obj/resourcesystem/images/062/0620a973-157a-4c90-b429-192d2db85fc3/4f65706bd495c883.png) |
| Out[1]= |
Get a list of five random SMILES:
| In[2]:= |
| Out[2]= | 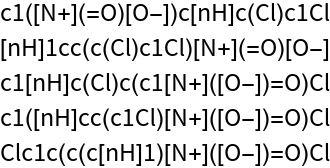 |
All of the strings encode the same molecular structure:
| In[3]:= |
| Out[3]= |
Compare this with the "IsomericSMILES" property, which lists the atoms in canonical order:
| In[4]:= |
| Out[4]= |
To get a reproducible SMILES string, use SeedRandom:
| In[5]:= |
| Out[5]= |
| In[6]:= | ![{SeedRandom[1234]; ResourceFunction["RandomSmilesString"]["caffeine"],
SeedRandom[1234]; ResourceFunction["RandomSmilesString"]["caffeine"]}](https://www.wolframcloud.com/obj/resourcesystem/images/062/0620a973-157a-4c90-b429-192d2db85fc3/254280d0ad9e732e.png) |
| Out[6]= |
Use BlockRandom to block one use of RandomSmilesString from affecting others:
| In[7]:= |
| Out[7]= |
By default single and aromatic bonds are elided. Use "AllBondsExplicit"→True to change this:
| In[8]:= |
| Out[8]= |
By default implicit hydrogen counts are only included when necessary. Use "AllHsExplicit"→True to change this:
| In[9]:= |
| Out[9]= |
Use "Kekulized"→True to use localized single and double bonds in place of aromatic bonds:
| In[11]:= |
| Out[11]= |
Use IncludeHydrogens→True to include hydrogen atoms as explicit atoms:
| In[12]:= |
| Out[12]= |
Small molecules will have a limited number of SMILES representations:
| In[13]:= |
| Out[13]= |
| In[14]:= |
| Out[14]= |
This work is licensed under a Creative Commons Attribution 4.0 International License