Resource retrieval
Get the pre-trained net:
Basic usage
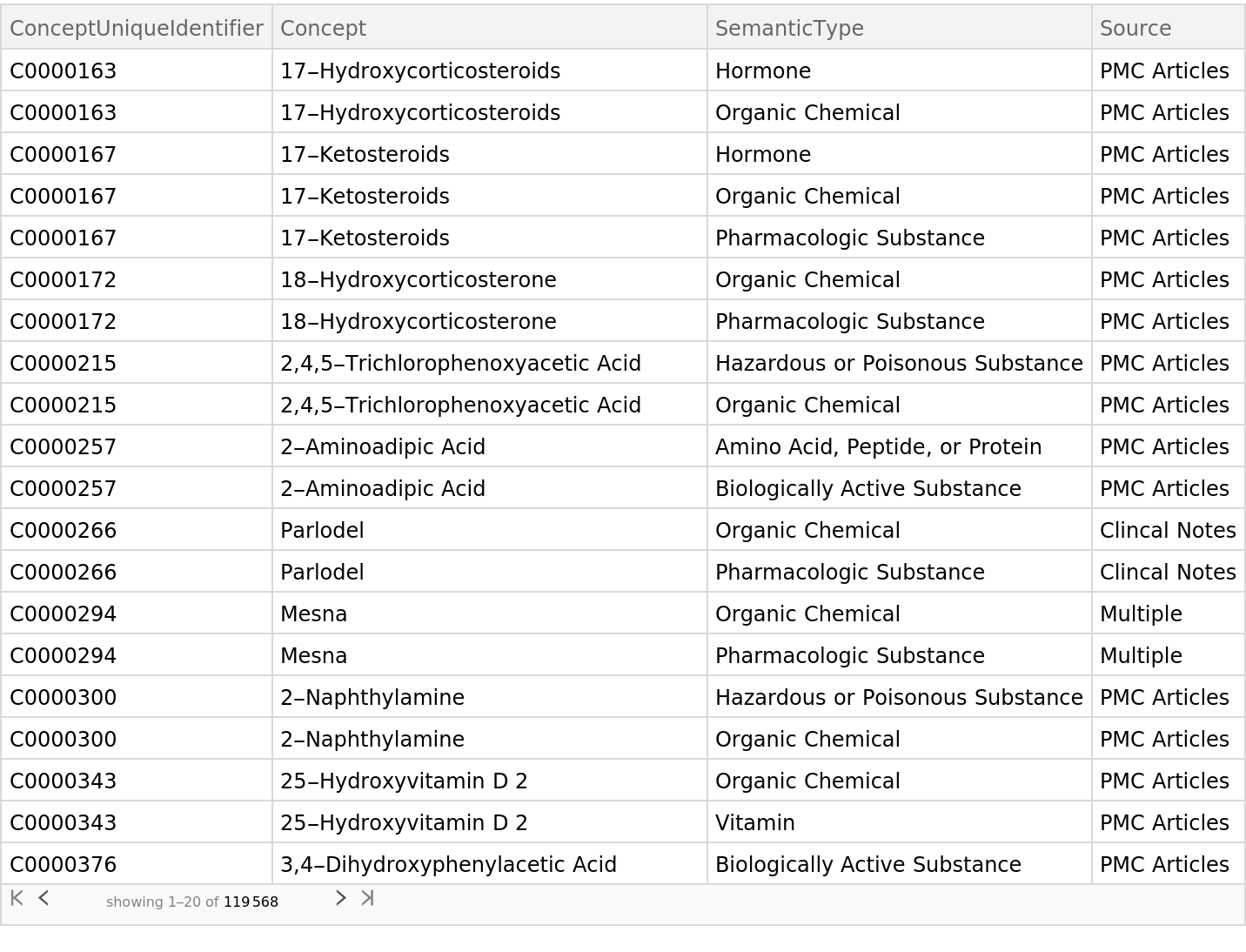
This net represents clinical concepts as 500-dimensional vectors. Concepts are identified by their concept unique identifier (CUI). The mapping between identifiers and human-readable concepts can be obtained from the clinical concepts ResourceData:
Find the concept unique identifier for the clinical concept "apnea":
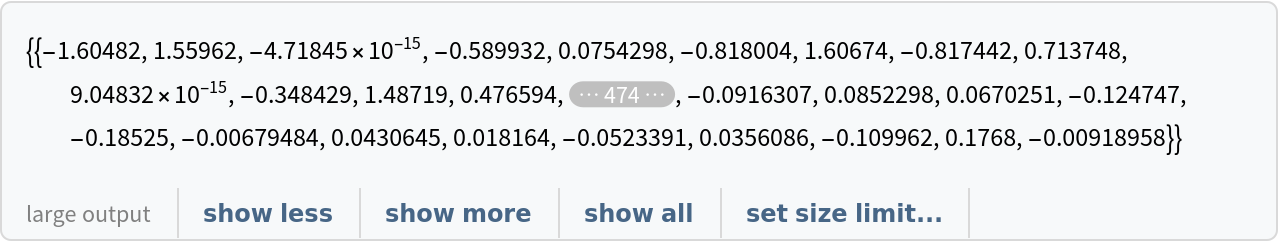
Use the net to obtain the associated embedded vector:
Use the embedding layer inside a NetChain:
Feature visualization
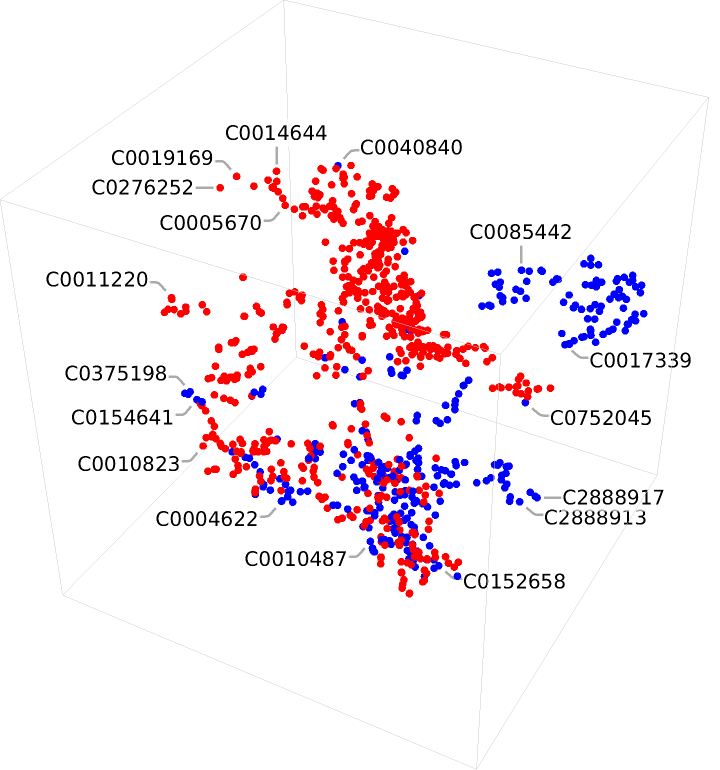
Create two lists of related concepts:
Visualize the associated embeddings in two dimensions:
Visualize the associated embeddings in three dimensions:
Word analogies
Get the pre-trained net:
Get a list of concept unique identifiers:
Obtain the embeddings:
Create an association whose keys are concept unique identifiers and whose values are vectors:
Find the concept unique identifier for the clinical concept "DNA virus":
Find the five nearest concept unique identifiers to "DNA virus":
Obtain the human-readable concept labels for these concept unique identifiers:
Explore similar drugs to a given one. Find the concept unique identifier for "metronidazole":
Find the five nearest concept unique identifiers to "metronidazole":
Obtain the human-readable concept labels for these concept unique identifiers:
Identify comorbidity relationships: a comorbidity is a disease or condition that frequently accompanies the primary diagnosis. A comorbidity for the condition "premature infant" is "bronchopulmonary dysplasia." Comorbidities of another condition--for example, obesity--can be investigated using word analogies. First obtain the relevant CUIs:
"Premature infant" is to "bronchopulmonary dysplasia" as "obesity" is to:
Obtain the human-readable concept labels for these concept unique identifiers:
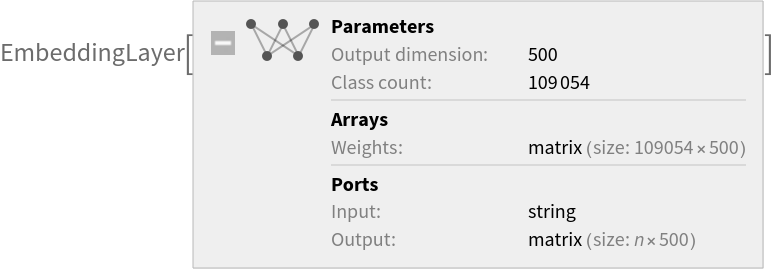
Net information
Inspect the sizes of all arrays in the net:
Obtain the total number of parameters:
Obtain the layer type counts:
Export to MXNet
Export the net into a format that can be opened in MXNet:
Export also creates a net.params file containing parameters:
Get the size of the parameter file:
The size is similar to the byte count of the resource object:
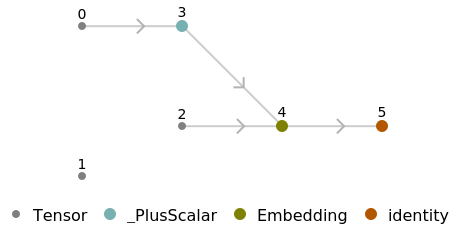
Represent the MXNet net as a graph:
![NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/137a8da9695a7688.png)
![apnea = First[
Normal[ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[StringMatchQ[#["Concept"], "apnea", IgnoreCase -> True] &],
"ConceptUniqueIdentifier"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/285390cd862e8689.png)
![vector = NetModel[
"Clinical Concept Embeddings Trained on Health Insurance Claims, \
Clinical Narratives from Stanford and PubMed Journal Articles"][apnea]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/0b194bc34e07dbba.png)
![chain = NetChain[{NetModel[
"Clinical Concept Embeddings Trained on Health Insurance Claims, \
Clinical Narratives from Stanford and PubMed Journal Articles"], LongShortTermMemoryLayer[10]}]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/41a8edd78fc5f373.png)

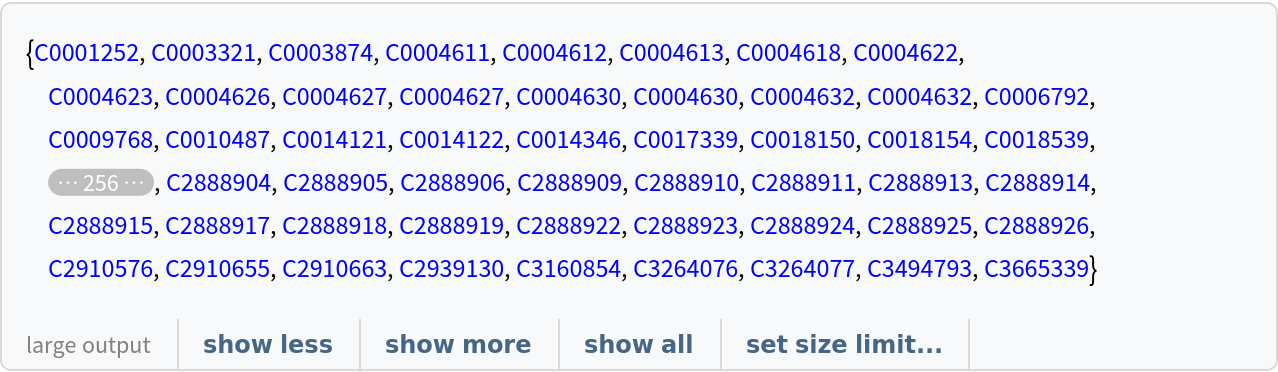
![viruses = Style[#, Red] & /@ Normal[ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[StringContainsQ[#["Concept"], "virus", IgnoreCase -> True] &], "ConceptUniqueIdentifier"]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/7f77a811833a6061.png)
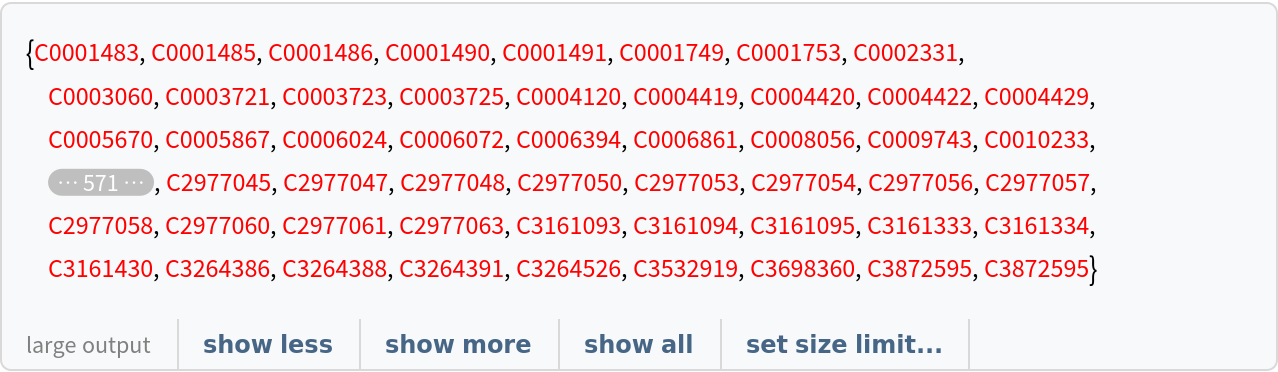
![bacteria = Style[#, Blue] & /@ Normal[ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[StringContainsQ[#["Concept"], "bacteria", IgnoreCase -> True] &], "ConceptUniqueIdentifier"]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/77dcd53587af9bcb.png)
![FeatureSpacePlot[Join[bacteria, viruses], FeatureExtractor -> NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/3a84a066c6ab926b.png)

![FeatureSpacePlot3D[Join[bacteria, viruses], FeatureExtractor -> NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/369fb55629bc140c.png)
![net = NetModel[
"Clinical Concept Embeddings Trained on Health Insurance Claims, \
Clinical Narratives from Stanford and PubMed Journal Articles"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/14169b89c4aeebca.png)
![dnavirus = First[Normal[
ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[StringContainsQ[#["Concept"], "DNA virus", IgnoreCase -> True] &], "ConceptUniqueIdentifier"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/5c71743ab322c487.png)

![metronidazole = First[Normal[
ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[StringContainsQ[#["Concept"], "metronidazole", IgnoreCase -> True] &], "ConceptUniqueIdentifier"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/67b438efa8bc4e3b.png)

![{premature, dysplasia, obesity} = Table[First@
Normal@ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[#Concept == concept &], "ConceptUniqueIdentifier"], {concept, {"Infant, Premature", "Bronchopulmonary Dysplasia", "Obesity"}}]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/0fb183b215c47fd4.png)
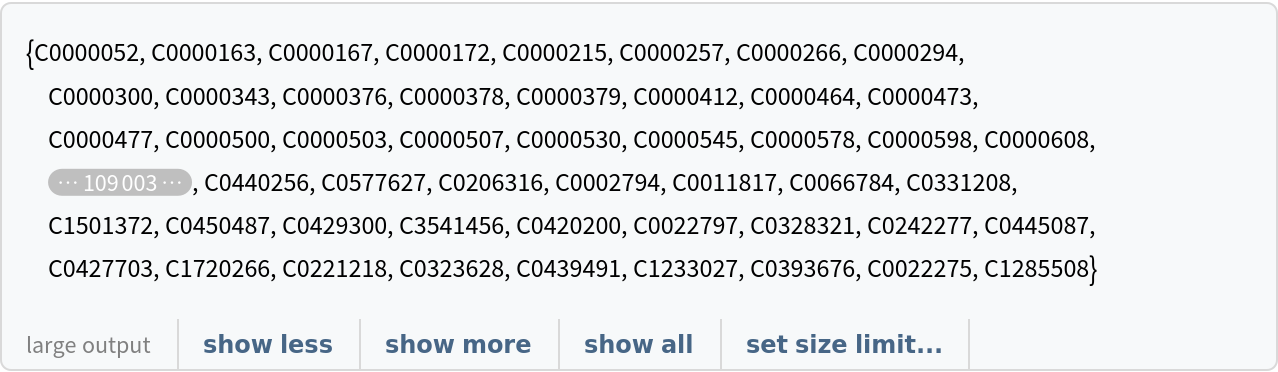
![ResourceData[
"Clinical Concepts from Massive Sources of Medical Data"][
Select[StringContainsQ[#["ConceptUniqueIdentifier"], results, IgnoreCase -> True] &], "Concept"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/7cabca8cc890abdd.png)

![NetInformation[
NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"], "ArraysElementCounts"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/566c746196ad0efe.png)
![NetInformation[
NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"], "ArraysTotalElementCount"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/6a503b106f910424.png)
![NetInformation[
NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"], "LayerTypeCounts"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/4ba643d5fa93bebd.png)
![jsonPath = Export[FileNameJoin[{$TemporaryDirectory, "net.json"}], NetModel["Clinical Concept Embeddings Trained on Health Insurance \
Claims, Clinical Narratives from Stanford and PubMed Journal \
Articles"], "MXNet"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/6261fb6e278e6cfc.png)
![ResourceObject[
"Clinical Concept Embeddings Trained on Health Insurance Claims, \
Clinical Narratives from Stanford and PubMed Journal \
Articles"]["ByteCount"]](https://www.wolframcloud.com/obj/resourcesystem/images/521/52127fd1-69f2-4b8e-b969-6c2099937396/2ef94161a55830ff.png)