U-Net
Trained on
Glioblastoma-Astrocytoma U373 Cells on a Polyacrylamide Substrate Data
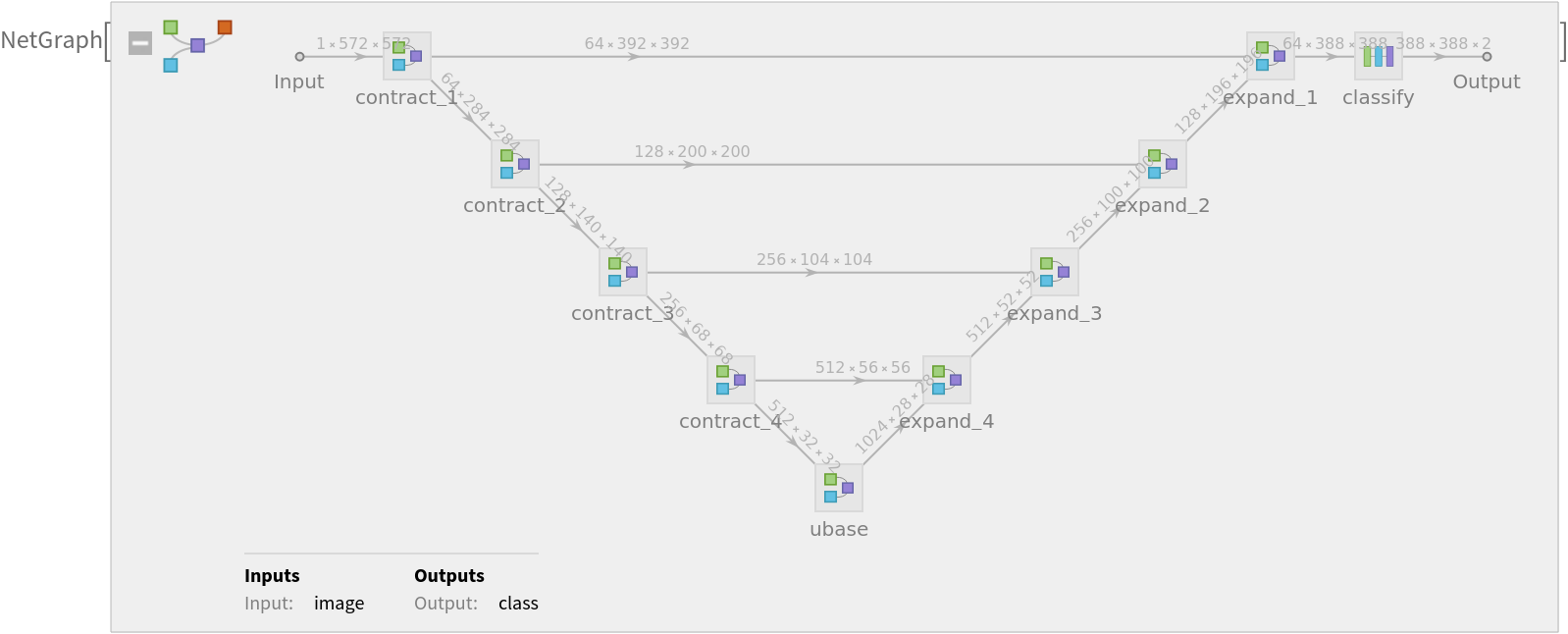
Released in 2015 by the University of Freiburg, Germany, this model exploits an architecture consisting of a contracting path to capture context and a symmetric expanding path that enables the precise segmentation of glioblastoma-astrocytoma U373 cells on a polyacrylamide substrate.
Number of layers: 61 |
Parameter count: 31,100,354 |
Trained size: 125 MB |
Examples
Resource retrieval
Get the pre-trained net:
Evaluation function
Define an evaluation function to handle net reshaping and tiling of the input and output:
Basic usage
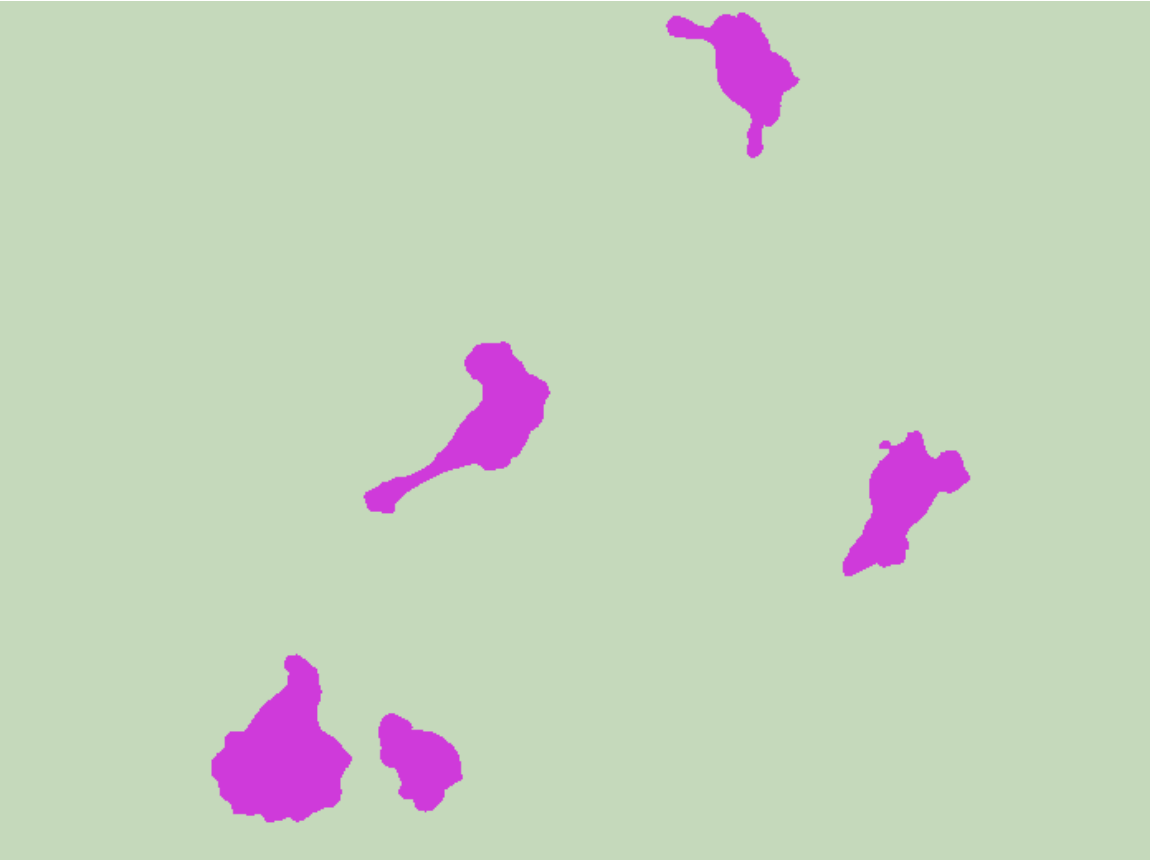
Obtain the segmentation mask for a given image:
Visualize the mask:
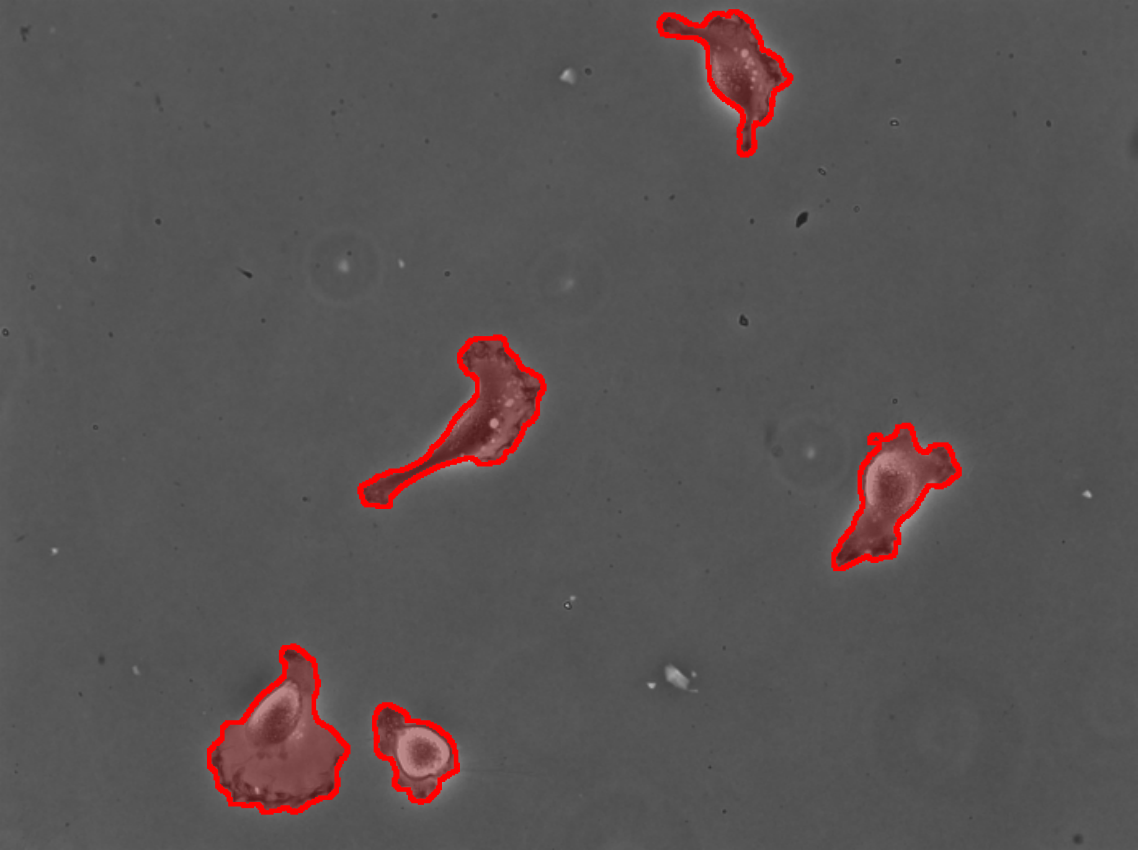
Overlay the mask on the input image:
Net information
Inspect the number of parameters of all arrays in the net:
Obtain the total number of parameters:
Obtain the layer type counts:
Display the summary graphic:
Export to MXNet
Export the net into a format that can be opened in MXNet:
Export also creates a net.params file containing parameters:
Get the size of the parameter file:
The size is similar to the byte count of the resource object:
Requirements
Wolfram Language
11.3
(March 2018)
or above
Resource History
Reference
-
O. Ronneberger, P. Fischer, T. Brox "U-Net: Convolutional Networks for Biomedical Image Segmentation," arXiv:1505.04597 (2015)
- Available from: https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/index.html
-
Rights:
All code is provided for research purposes only and without any warranty. Any commercial use requires the authors' consent. When using the code in your research work, you should cite the respective paper.
![netevaluate[img_, device_ : "CPU"] :=
Block[
{net = NetModel[
"U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"],
dims = ImageDimensions[img], pads, mask},
pads = Map[{Floor[#], Ceiling[#]} &, Mod[4 - dims, 16]/2];
mask = NetReplacePart[
net,
{"Input" -> NetEncoder[{"Image", Ceiling[dims - 4, 16] + 188, ColorSpace -> "Grayscale"}],
"Output" -> NetDecoder[{"Class", Range[2], "InputDepth" -> 3}]}][
ImagePad[ColorConvert[img, "Grayscale"], pads + 92, Padding -> "Reversed"],
TargetDevice -> device
];
Take[mask, {1, -1} Reverse[pads[[2]] + 1], {1, -1} (pads[[1]] + 1)]
]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/7b13d949beff3f76.png)
![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/2fd8477a-208c-4858-b691-ef930a652612"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/26710e6ceb4ff350.png)
![NetInformation[
NetModel["U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"], "ArraysElementCounts"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/44bf328ed9efb753.png)

![NetInformation[
NetModel["U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"], "ArraysTotalElementCount"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/05d21f0268909156.png)
![NetInformation[
NetModel["U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"], "LayerTypeCounts"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/5cb5cec2ffe077ac.png)
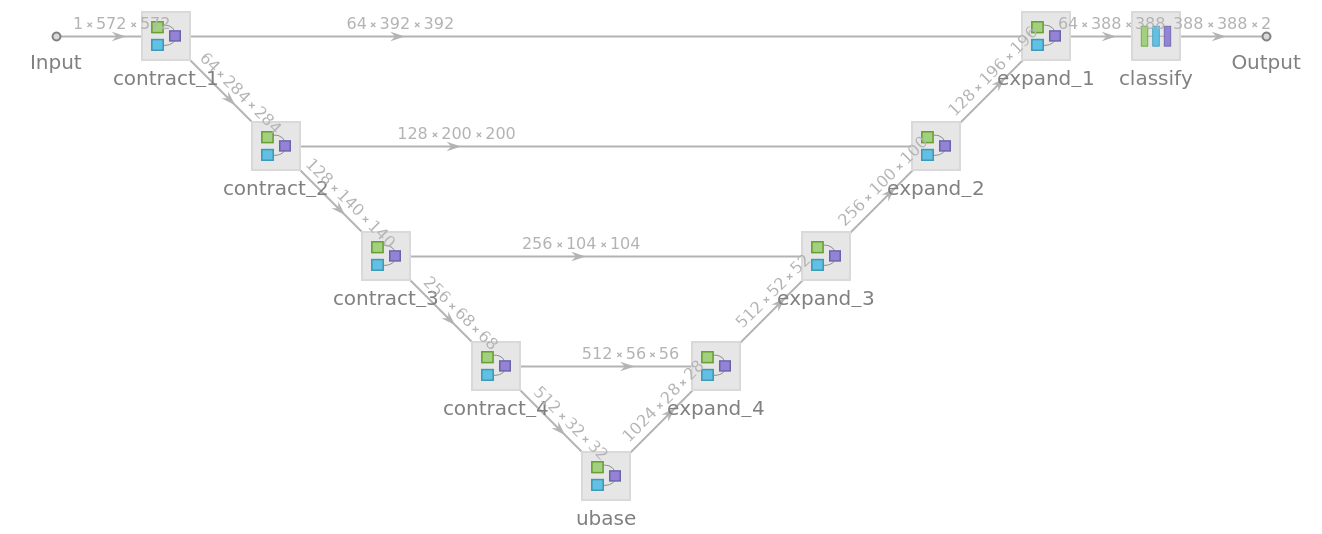
![NetInformation[
NetModel["U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"], "SummaryGraphic"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/5a4cb943b0fc3791.png)
![jsonPath = Export[FileNameJoin[{$TemporaryDirectory, "net.json"}], NetModel["U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"], "MXNet"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/793cc14ea45cb0a7.png)
![ResourceObject[
"U-Net Trained on Glioblastoma-Astrocytoma U373 Cells on a \
Polyacrylamide Substrate Data"]["ByteCount"]](https://www.wolframcloud.com/obj/resourcesystem/images/142/14214182-07c2-4c00-a65a-fbc45066ebba/7351a7ae270b09c7.png)