ForkNet Brain Segmentation Net
Trained on
NAMIC Data
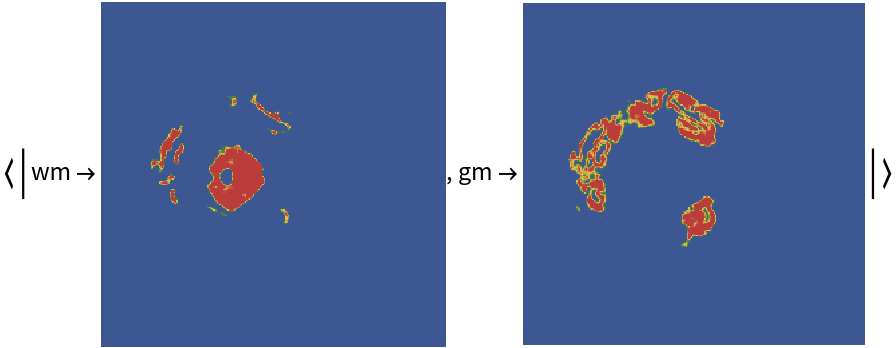
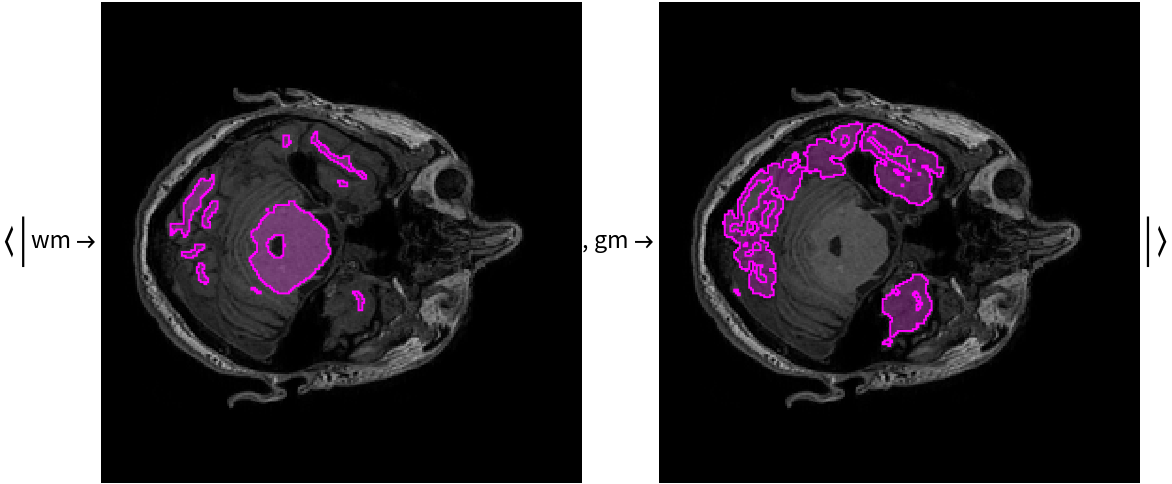
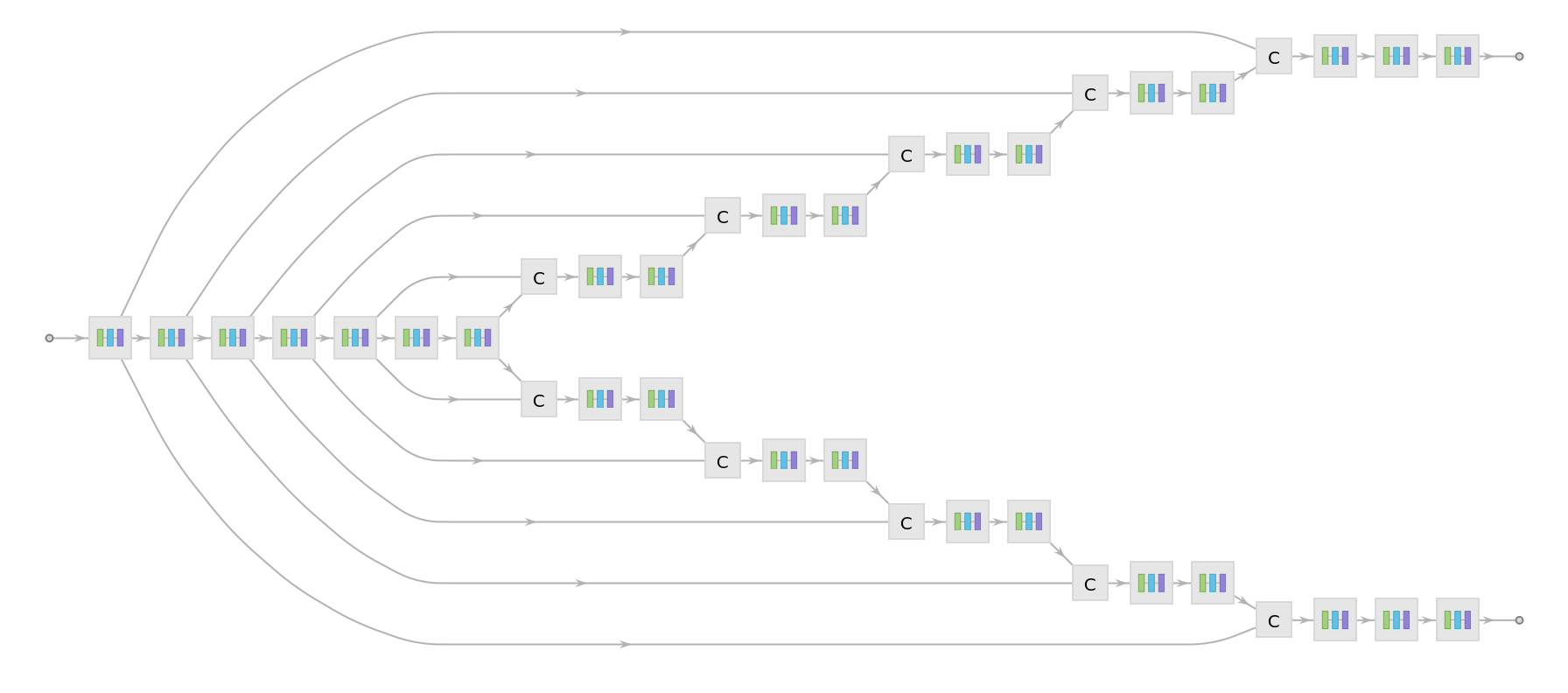
Released in 2020 by a research team from Nagoya Institute of Technology (Japan), ForkNet Segmentation Net is a u-net inspired CNN that performs segmentation of various brain tissues. The model is comprised of several u-net structures with shared encoders and distinct decoders corresponding to distinct brain tissues. The segmented tissues can be used to generate personalized head models and applied for the evaluation of the electric field in the brain during transcranial magnetic stimulation. Compared to vanilla u-net architecture, the networks get substantial gain in accuracy once the number of decoders is increased.
Number of models: 5
Examples
Resource retrieval
Get the pre-trained net:
NetModel parameters
This model consists of a family of individual nets, each identified by a specific parameter combination. Inspect the available parameters:
Pick a non-default net by specifying the parameters:
Pick a non-default uninitialized net:
Basic usage
Obtain the default segmentation masks for a given image:
Obtain the dimensions of the masks:
Visualize the masks:
Overlay the masks on the input image:
Net information
Inspect the number of parameters of all arrays in the net:
Obtain the total number of parameters:
Obtain the layer type counts:
Display the summary graphic:
Export to MXNet
Export the net into a format that can be opened in MXNet:
Export also creates a net.params file containing parameters:
Get the size of the parameter file:
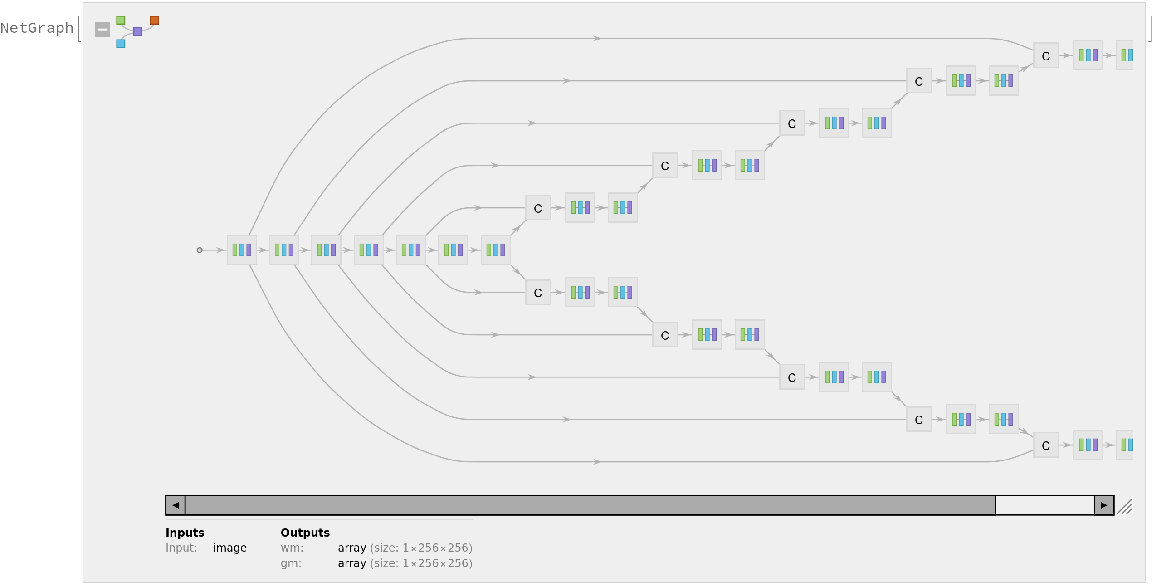
Represent the MXNet net as a graph:
Requirements
Wolfram Language
12.1
(March 2020)
or above
Resource History
Reference
-
E.-A. Rashed, J. Gomez-Tames, A. Hirata, "Development of Accurate Human Head Models for Personalized Electromagnetic Dosimetry Using Deep Learning," Neuroimage, 202, 116132 (2019)
E.-A. Rashed, J. Gomez-Tames, A. Hirata, "End-to-End Semantic Segmentation of Personalized Deep Brain Structures for Non-invasive Brain Stimulation," Neural Networks, 125, 233–244 (2020)
- Available from:
-
Rights:


![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/f2456b07-2f56-4614-b2fb-41ab4b010e8c"]](https://www.wolframcloud.com/obj/resourcesystem/images/01c/01c69db4-af3c-48cf-a425-8bd455b2855c/173ccf5c98796a81.png)